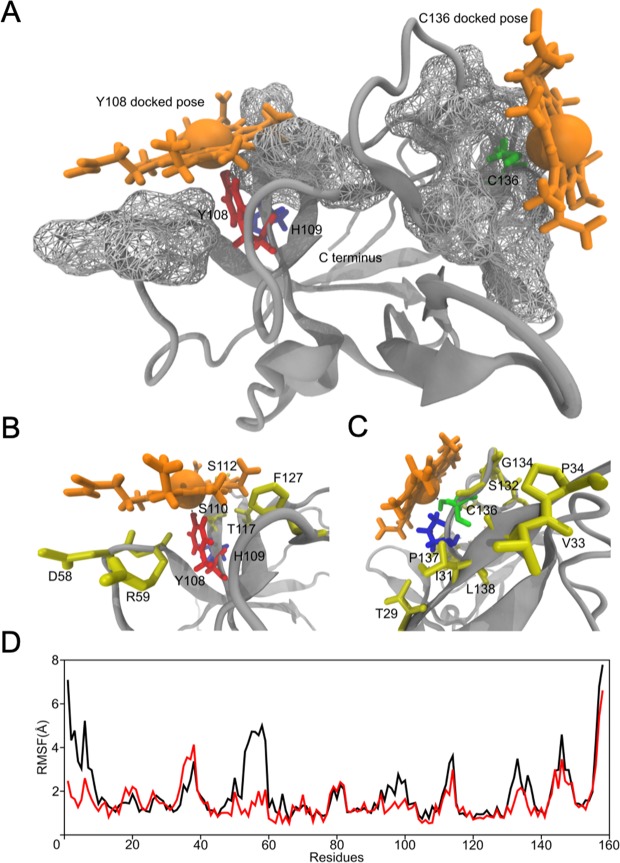
Figure 4.
Heme binding to IL-36α and its effects on the protein supported by molecular docking and MD simulation studies. (A–C) The structure of wild-type IL-36α (grey) with two heme molecules (orange) docked at C136 (green) and Y108 (red), respectively (H109, blue). Wireframe surfaces drawn around both binding sites (C136, Y108) represent surfaces of IL-36α residues that make contact with the heme molecule as predicted by the docking algorithm (A). A zoom in at Y108 (B, Y108, red; H109, blue) and C136 (C, C136, green; P137, blue). Residues that stabilize the heme-bound conformation (yellow) are labeled. (D) Comparative per-residue root mean squared fluctuation (RMSF) profiles of the heme bound (red) and the free states (black) of the protein. The RMSF profiles were generated from 200 ns equilibration MD simulations of the free protein and the protein-heme complex. Clear reduction of fluctuations from their mean position is noticed for several amino acids in the heme-bound state indicating the effect of heme binding to the protein.