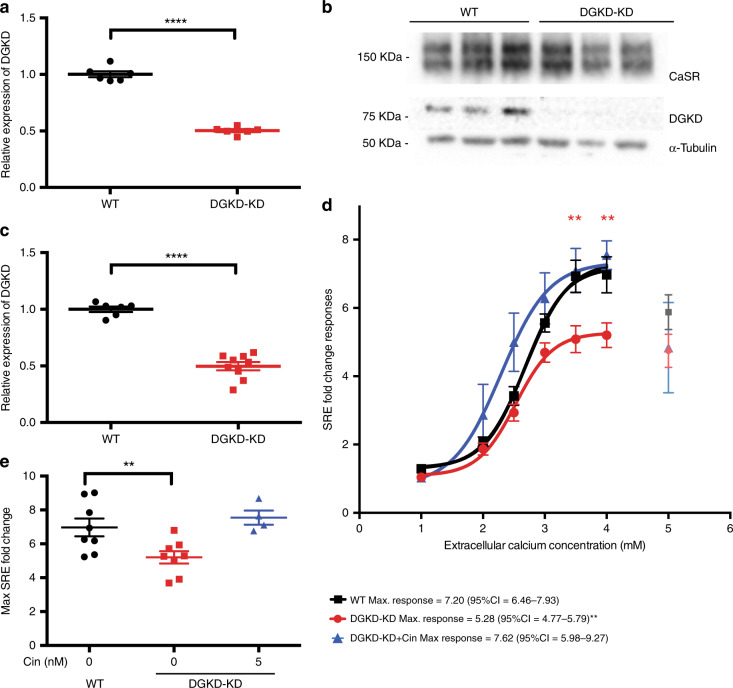
Fig. 3.
CaSR-mediated SRE responses following DGKD knockdown and effect of cinacalcet treatment in HEK-CaSR-SRE cells. a Relative expression of DGKD, as assessed by quantitative real-time PCR of HEK-CaSR-SRE cells treated with scrambled (WT) or DGKD (DGKD-KD) siRNA and used for SRE experiments. Samples were normalized to a geometric mean of four housekeeper genes: PGK1, GAPDH, TUB1A, CDNK1B. n = 8 biologically independent transfections. b Representative western blot of lysates from HEK-CaSR cells treated with scrambled or DGKD siRNA and used for SRE experiments. α−Tubulin was used as a loading control. c Relative expression of DGKD, as assessed by densitometry of western blots from cells treated with scrambled or DGKD siRNA demonstrating a ~50% reduction in expression of DGKD following treatment with DGKD siRNA. Samples were normalized to PGK1. n = 6 biologically independent transfections for WT, n = 9 biologically independent transfections for DGKD-KD. d SRE responses of HEK-CaSR-SRE cells in response to changes in extracellular calcium concentration. Cells were treated with scrambled (WT) or DGKD (DGKD-KD) siRNA. The responses ± SEM are shown for n = 8 biologically independent transfections for WT and DGKD-KD cells and n = 4 biologically independent transfections for DGKD-KD + 5 nM cinacalcet cells. Treatment with DGKD siRNA led to a reduction in maximal response (red line) compared to cells treated with scrambled siRNA (black line). This loss-of-function could be rectified by treatment with 5 nM cinacalcet (blue line). Post desensitization points are shown but were not included in the analysis (gray, light red, and light blue). e Mean maximal responses with SEM of cells treated with scrambled siRNA (WT, black), DGKD siRNA (DGKD-KD, red) and DGKD siRNA incubated with 5 nM cinacalcet (blue). Statistical comparisons of maximal response were undertaken using F test. Student’s t-tests were used to compare relative expression. Two-way ANOVA was used to compare points on dose response curve with reference to WT. Data are shown as mean ± SEM with **p < 0.01, ****p < 0.0001. Source data are provided as a Source Data file