Figure 1. HyCoSuL screening of Lon substrates.
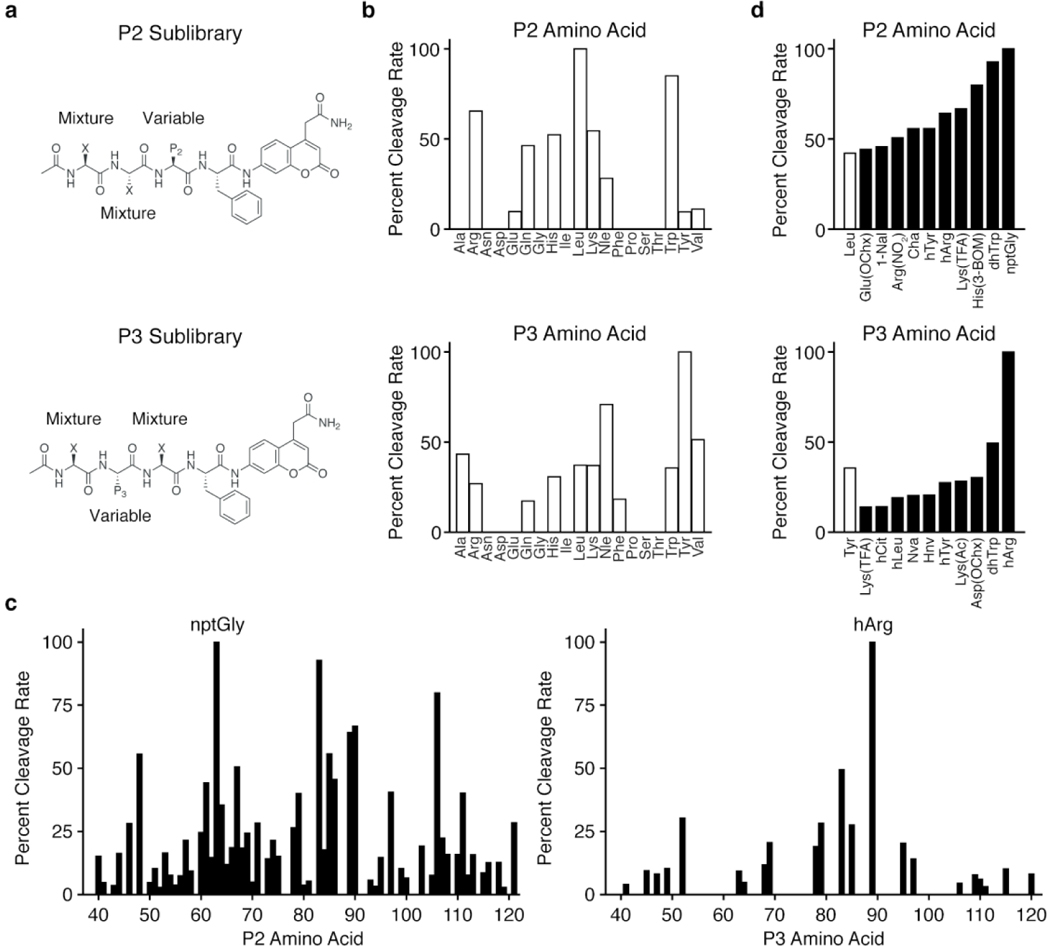
Structures of (a) P2 and P3 fluorogenic HyCoSuL libraries. Positions comprising equimolar mixtures of 18 natural amino acids and norleucine (Nle) as a substitute for Met and Cys (not included in the library) are designated by an X. The remaining variable position is held constant as the indicated natural or non-natural amino acid for each sub-library. Plot of the relative cleavage rates for (b) natural amino acids in the P2 (top) or P3 (bottom) positions and (c) non-natural amino acids in the P2 (left) or P3 (right) positions. (d) Plots of cleavage rates of the best non-natural amino acids for the P2 (top) or P3 (bottom) positions. The best natural amino acid (white bar) at each position is included for comparison. Results for each sub-library were normalized to the amino acid with the fastest cleavage rate, indicated by an arrow, and data represent the mean of two independent screens. Amino acid type is indicated by white (natural) or black (non-natural) bars. D-amino acids (Nos. 20-36) exhibited no cleavage and were excluded from the plot in (c). Amino acid abbreviations and structures are as reported in Kasperkiewicz, et al37. The list of amino acids and normalized cleavage rates can be found in Table S1.
