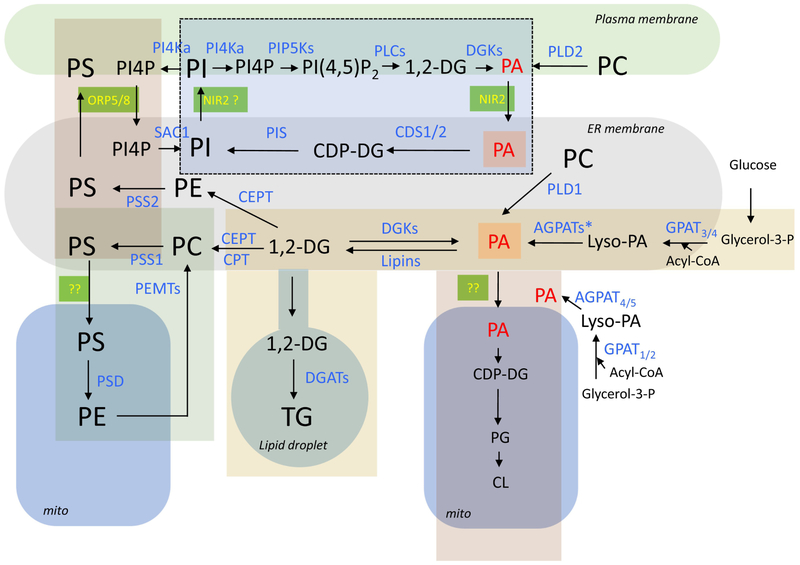
Figure 1:
Enzymatic pathways of lipid synthesis and their membrane localization in a generic mammalian cell. The enzymes are labeled blue and the lipids are black except PA (red) which represents a major hub for several metabolic pathways. Major organelles are designated by the rounded shapes. Metabolic units that have been linked are designated by rectangles of different colors. The dotted lines denote inositol lipids that serve as signaling entities. Lipid transfer proteins working in contact sites are labeled yellow on green background. Note that lipids in the ER can be part of multiple metabolic units and how these are kept separated is one of the important questions yet to be understood. Abbreviations of enzymes: PI4K, PI 4-kinase; PIP5K, PI4P 5-kinase; PLC, phospholipase C; DGK, diacylglycerol kinase; PLD, phospholipase D; Sac1, Sac1 phosphatase; PIS, phosphatidylinositol synthase; CDS, CDP-DG synthase; PSS, phosphatidylserine synthase; CEPT, choline/ethanolamine phosphotransferase; CPT, CDP-choline:1,2-diacylglycerolphosphocholine transferase; PSD, phosphatidylserine decarboxylase; PEMT, Phosphatidylethanolamine N-methyltransferase; DGAT, diacylglycerol acyl transferase; AGPATs, 1-Acylglycerol-3-Phosphate O-Acyltransferase (* denotes that several forms of these enzymes exist in the ER. AGPAT4 and −5 are found in mitochondria); GPAT, Glycerol-3-Phosphate Acyltransferase. Abbreviation of lipids: PI, phosphatidylinositol; PI4P, phosphatidylinositol 4-phosphate; PI(4,5)P2, phosphatidylinositol 4,5-bisphosphate; DG, diacylglycerol; PA, phosphatidic acid; PS, phosphatidylserine; PC, phosphatidylcholine; PE, phosphatidylethanolamine; TG, triacylglycerol; PG, phosphatidylglycerol; CL, cardiolipin; CDP-DG, cytidine-diphosphate-diacylglycerol.