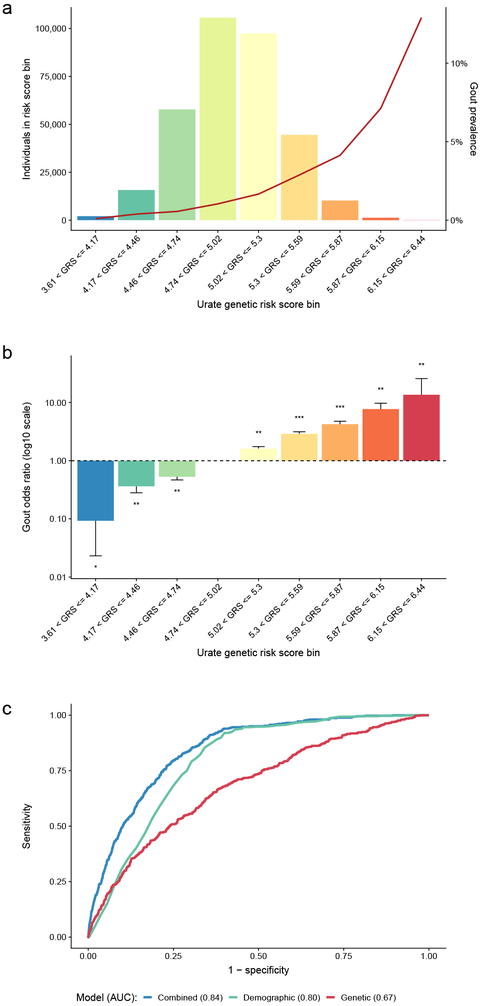
Figure 2 ∣. A genetic risk score (GRS) for serum urate improves gout risk prediction.
a, Histogram of the urate GRS among 334,880 European ancestry participants of the UK Biobank. The y-axes show the number of individuals (left) and the prevalence of gout (right), the x-axis shows categories of the urate GRS. The units on the x-axis represent genetically predicted serum urate levels (mg/dl) compared to individuals without any urate-increasing alleles. b, Age- and sex-adjusted odds ratio of gout (y-axis) by GRS category (x-axis) among 334,880 European-ancestry participants of the UK Biobank, comparing each category to the most prevalent category (4.74 < GRS ≤ 5.02) with error bars representing 95% confidence intervals; * denotes logistic regression two-sided P-value < 0.05, ** denotes P < 5 × 10−10, and *** P < 5 × 10−100. c, Comparison of the receiver operating characteristic (ROC) curves of different prediction models of gout: genetic (GRS only; red), demographic (age + sex; green), and combined (GRS + age + sex; blue). y-axis: sensitivity, x-axis: specificity. At the optimal cut points determined by the maximum of the Youden’s index, the sensitivity of the combined model was 84% and specificity was 68%.