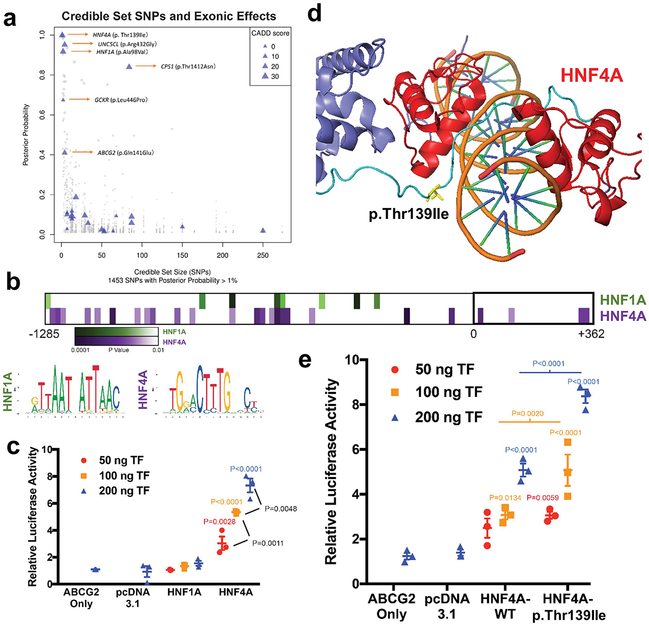
Figure 5 ∣. Prioritization of p.Thr139Ile at HNF4A and functional study of HNF4A regulation of ABCG2 transcription.
a, Graph shows credible set size (x-axis) against the posterior probability of association (PPA; y-axis) for each of 1,453 SNPs with PPA > 1% in 114 99% credible sets. Triangles mark missense SNPs, with size proportional to their Combined Annotation Dependent Depletion (CADD) score. Blue triangles indicate missense variants mapping into small (≤ 5 SNPs) credible sets or with high PPA (≥ 50%). b, Predicted HNF1A or HNF4A binding sites in the promoter region of ABCG2 using LASAGNA 2.0, the consensus affinity sequence, and the P-value of likely matches based on nucleotide position within a consensus transcription factor binding site (Methods). c, Relative luciferase activity and transactivation of ABCG2 promoter in cells transfected with variable amount of HNF1A or HNF4A constructs (mean (line) ± s.e.m. (whiskers), n = 3 independent experiments, P-values calculated with ordinary one-way ANOVA with Tukey’s multiple comparison test). d, Position of p.Thr139Ile (T139I) in DNA binding domain/hinge region within HNF4A homodimer structure (PDB 4IQR). e, Relative luciferase activity and transactivation of ABCG2 promoter in cells transfected with variable amount of constructs (ng’s of transfected DNA) of wild-type HNF4A (threonine) or isoleucine at position 139 (± s.e.m., n = 3 independent experiments, P-values calculated with ordinary one-way ANOVA with Tukey’s multiple comparison test).