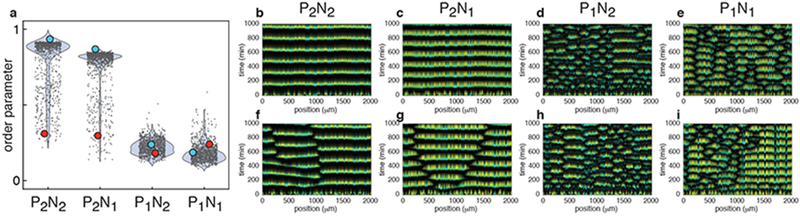
Figure 3.
The mathematical model reproduces the experimentally observed spatio-temporal dynamics exhibited by the four regulatory architectures. (a) Calculated order parameters for different initial conditions. The width of the shaded blue regions represents the empirical probability density of the distribution of the individual results (dots). The same set of 103 random initial phases and ratios between two strains were used in simulations of all four architectures (n=103 independent simulations for each). The order parameters for the P2N1 and P2N1 architectures were much higher than those for the P1N2 and P1N1 architectures. (b-e) Representative simulations using the same initial conditions (blue dots in (a)) that resulted in high order parameters in the P2N2 and P2N1 architectures, but not in the P1N2 and P1N1 architectures (n=103 independent simulations for each). (f-i) Representative simulations using the same initial conditions (red dots in (a)) that resulted in low order parameters in each regulatory architecture (n=103 independent simulations for each). Low order parameters in simulations with P2N1 and P2N1 architectures reflected the emergence of 2-3 subpopulations oscillating in anti-phase (f, g). In contrast, low order parameters reflected spatially disorganized behavior in P1N2 and P1N1 architectures (h, i).