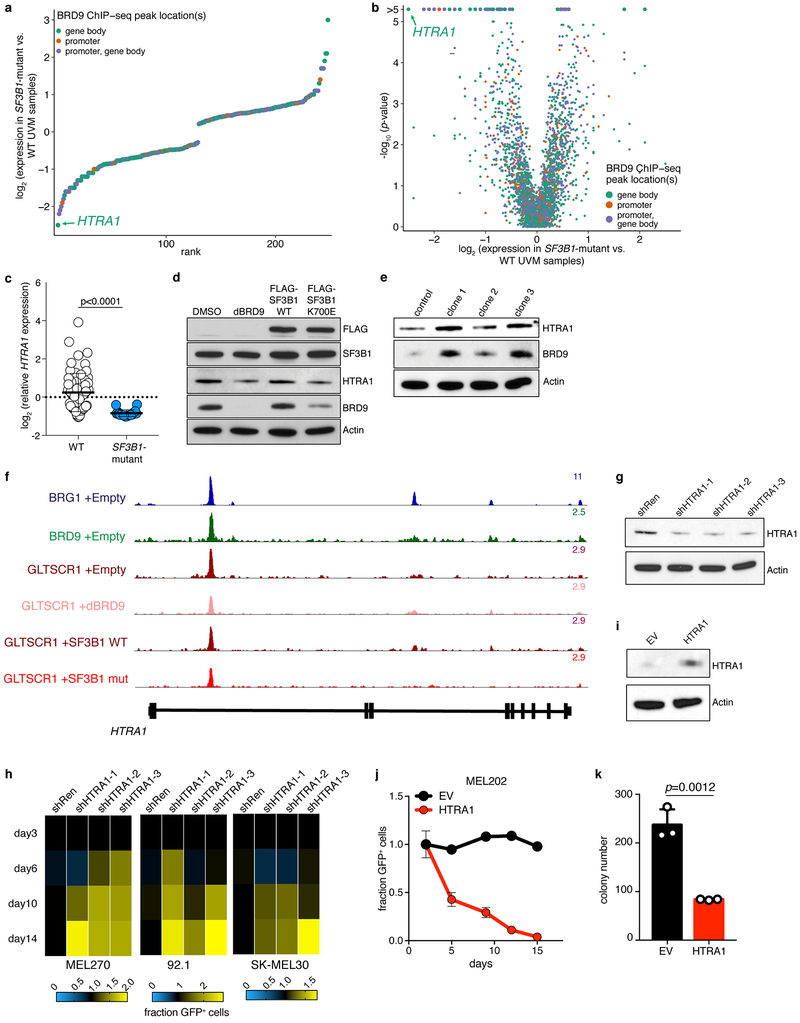
Extended Data Figure 7. BRD9 regulates HTRA1 expression to promote UVM tumorigenesis.
(a) Rank plot illustrating log fold-change of each significantly differentially expressed gene identified by comparing SF3B1-mutant to WT patient samples from the TCGA UVM cohort. Plot restricted to genes with BRD9 ChIP-seq peaks within their promoter or gene body in the absence of perturbations to BRD9 (MEL270 cells treated with DMSO or following ectopic expression of WT SF3B1). n=3,122 genes analyzed, of which n=248 met the significance (p < 0.001) and expression (median expression in both WT and mutant samples >2 transcripts per million) thresholds and so were illustrated here. p-value computed with a two-sided Mann-Whitney U test.
(b) As (a), but a volcano plot additionally illustrating the p-value associated with the comparison between SF3B1-mutant and WT samples. n=3,122 genes analyzed and illustrated. p-value computed with two-sided Mann-Whitney U test.
(c) HTRA1 expression in TCGA UVM samples with (n=18) or without (n=62) SF3B1 mutations. Expression is z-score normalized across all samples. Data are presented as mean ± SD. p-value computed with two-sided t-test.
(d) Western blot for FLAG, SF3B1, HTRA1, BRD9, and actin in SF3B1-WT MEL270 cells treated with DMSO, dBRD9, FLAG-SF3B1 WT cDNA, or FLAG-SF3B1 K700E cDNA. Representative images from n=3 biologically independent experiments.
(e) Western blot for HTRA1, BRD9, and Actin in MEL202 cells (SF3B1R625G) following CRISPR/Cas9-mediated mutagenesis of the BRD9 poison exon (as shown in Extended Data Fig. 2o). Representative images from n=3 technically independent experiments.
(f) Read coverage for BRG1, BRD9, and GLTSCR1 ChIP-seq at the HTRA1 locus in MEL270 cells (d) treated with an empty vector, dBRD9, or SF3B1 WT or K700E cDNAs (n=1 ChIP-seq experiment performed for each condition).
(g) Western blot for HTRA1 and Actin in MEL270 cells treated with anti-HTRA1 shRNAs or a non-targeting control shRNA (shRen). Representative images from n=3 biologically independent experiments.
(h) Heat map summarizing the results of a competition assay to measure the effect of each indicated shRNA on the growth of Cas9-expressing SF3B1-WT UVM cell lines. Cell growth was computed with respect to cells treated with a non-targeting control shRNA (shRen) and the percentages of GFP+ cells at day 14 was normalized to that at day 2. The illustrated values correspond to the mean computed over n=3 biologically independent experiments.
(i) Western blot for HTRA1 and actin in MEL202 cells (SF3B1R625G) following stable overexpression of an empty vector (EV) or HTRA1 (both in an MSCV-IRES-GFP vector). Representative images from n=3 biologically independent experiments.
(j) Ratio of GFP+ to GFP- MEL202 cells (SF3B1R625G) from a competition experiment in which GFP+ cells from (i) were seeded at an initial 1:1 ratio with GFP- control cells. Data are presented as mean of n=2 biologically independent experiments.
(k) Colony number of MEL202 cells expressing EV or HTRA1 cDNA from (i) following 10 days of growth in soft agar. Data are presented as mean of n=3 biologically independent experiments.