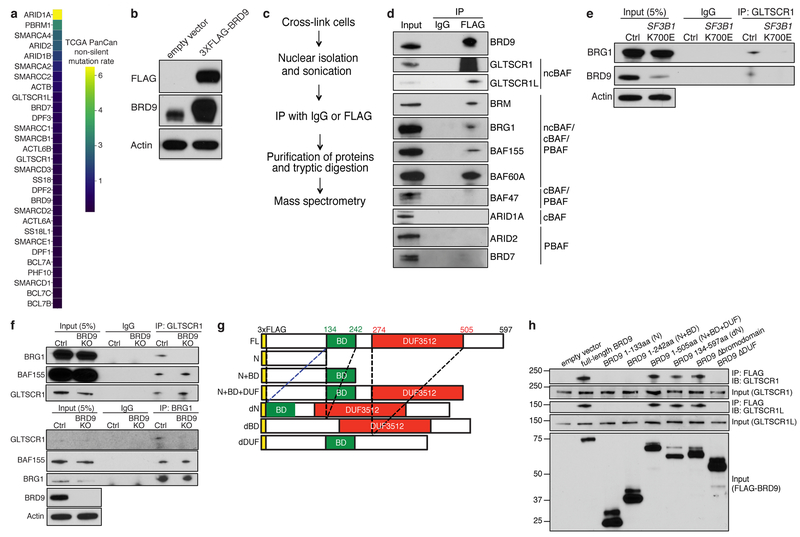
Extended Data Figure 3. BRD9 loss impairs non-canonical BAF complex formation.
(a) Mutation rate observed across TCGA cohorts for cBAF, PBAF, and ncBAF components.
(b) Western blot confirming FLAG-tagged BRD9 protein expression in 3XFLAG-BRD9-expressing K562 cells. Representative images from n=3 biologically independent experiments.
(c) Experimental workflow for using RIME (Rapid Immunoprecipitation Mass spectrometry of Endogenous proteins)32 for purification and identification of chromatin-associated interactions partners of BRD9.
(d) Cross-linking and immunoprecipitation with IgG or FLAG followed by probing with the indicated antibodies. Data from 3xFLAG-BRD9-expressing MEL270 cells. Representative images from n=3 biologically independent experiments.
(e) Immunoprecipitation of GLTSCR1 followed by Western blotting with the indicated antibodies in SF3B1K700E knockin NALM-6 cells. Representative images from n=3 biologically independent experiments.
(f) Immunoprecipitation of GLTSCR1 (top) or BRG1 (bottom) followed by blotting with the indicated antibodies in K562 cells with CRISPR-mediated knockout of BRD9. Representative images from n=3 biologically independent experiments.
(g) Schematic of the BRD9 full-length (FL) protein and deletion mutants constructed. BD, bromodomain; DUF, domain of unknown function. “EV”: empty vector; “FL”: full-length BRD9; “N”: 1-133 amino acids (aa) of BRD9; “N+BD”: 1-242aa of BRD9; “N+BD+DUF”: 1-505aa BRD9; “dN”: 134-597aa of BRD9; “dBD”: Bromodomain deletion mutant of BRD9; “dDUF”: DUF domain deletion mutant of BRD9.
(h) Immunoprecipitation (IP) with FLAG following by probing (IB) for GLTSCR1 or GLTSCR1L in 293T cells expressing 3xFLAG-tagged versions of the indicated deletion mutants. Deletion mutants illustrated in (g). Representative images from n=3 biologically independent experiments.