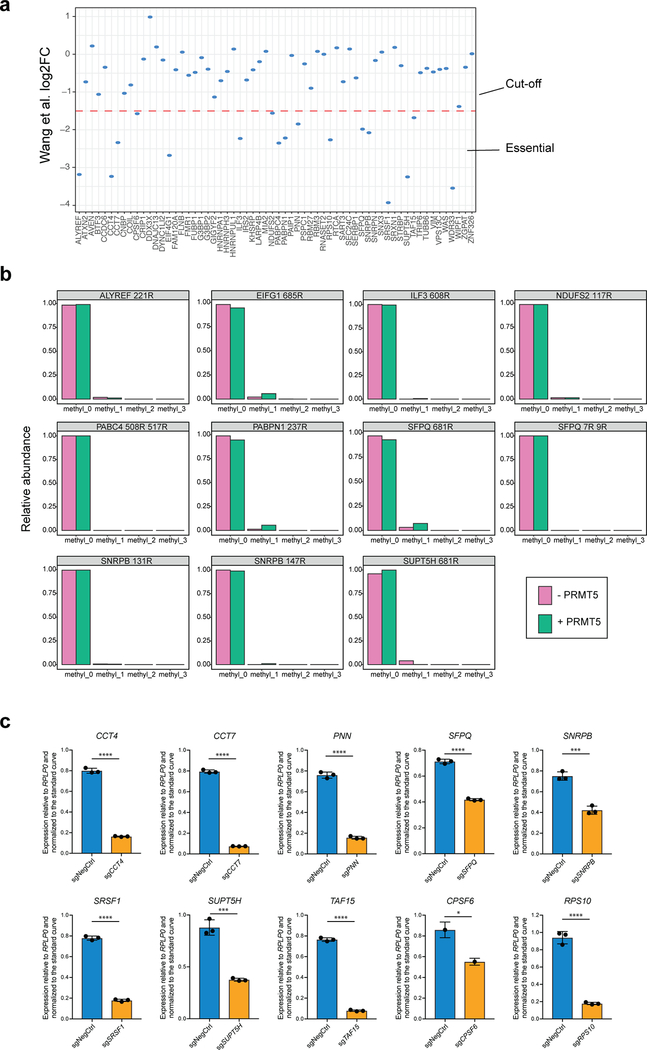
Extended Data Fig. 3. Validation of the essential PRMT5 substrates.
a, 17 out of 62 potential PRMT5 substrates were chosen as potentially essential according to a previously published CRISPRko screen in THP-1 cells. Y axis represents log2FC of the relative abundance of sgRNA in the screen and −1.5 was chosen as a cut-off. b, Distributions of relative abundances of unmethylated and methylated peptide forms after the incubation with or without recombinant PRMT5-WDR77 complex. Only the peptides belonging to the unconfirmed PRMT5 substrates are shown here. c, qRT-PCR analysis of CCT4, CC7, PNN, SFPQ, SNRPB, SRSF1, SUPT5H, TAF15, CPSF6 and RPS10 expression demonstrates efficient knockdown of the genes upon CRISPRi sgRNA transduction (n=3, * is p-value < 0.033, *** is p-value < 0.001, **** is p-value < 0.0001 according to the unpaired t test). The experiments were repeated twice with similar results.