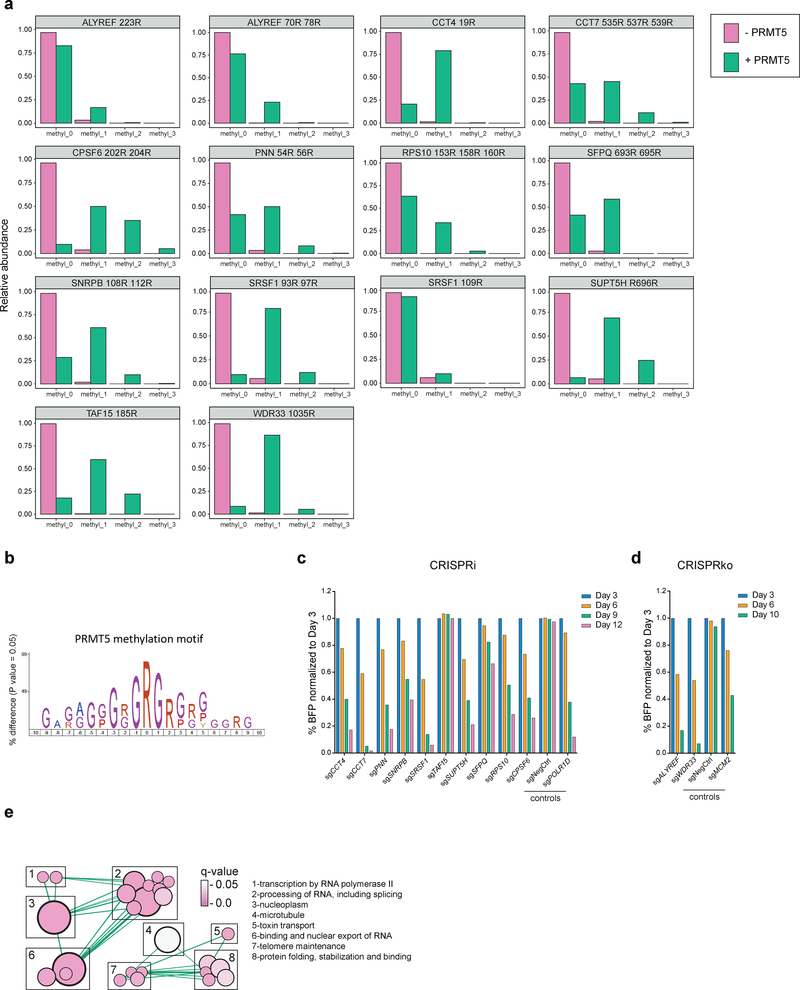
Figure 3. Validation of the essential PRMT5 substrates.
a, Distributions of abundances of unmethylated and methylated peptide forms after the incubation with or without recombinant PRMT5-WDR77 complex. Only the peptides belonging to the confirmed PRMT5 substrates are shown here. b, PRMT5 methylation motif predicted using an iceLogo tool. Y axis represents the difference between the frequency of an amino acid in a sample set and the reference set (human proteome). c, CRISPRi competition assays to confirm essentiality of CCT4, CCT7, PNN, SNRPB, SRSF1, TAF15, SUPT5H, SFPQ, RPS10 and CPSF6. THP-1-cdCas9-KRAB cells were transduced with the sgRNAs against the genes of interest and the percentage of sgRNA-transduced (BFP-positive) cells was measured over time. An sgRNA targeting POLR1D was used as a positive control and a non-targeting sgRNA (NegCtrl) was used as a negative control. d, CRISPRko competition assays to confirm essentiality of ALYREF and WDR33. THP-1-wtCas9 cells were transduced with lentiviruses expressing the sgRNAs against the genes of interest and the percentage of sgRNA-transduced (BFP-positive) cells was measured over time. An sgRNA targeting MCM2 was used as a positive control and a non-targeting sgRNA (NegCtrl) was used as a negative control. e, Enrichment map of the Gene Ontology-enriched protein sets across 11 validated essential substrates of PRMT5 (two-sided Fisher’s exact test, FDR-adjusted p-value < 0.05). Nodes represent significantly enriched protein sets, node size is proportional to the number of members in a protein set, and color intensity depends on the q-value. Edges indicate the protein overlap between the nodes with thicker edges indicating higher overlap between the nodes. Functionally related protein sets are clustered, numbered and named. The experiments in a, c, d were repeated twice with similar results. Source data for a are available in Supplementary Table 1. Source data for c,d are available online.