Abstract
Gene duplication is a common and powerful mechanism by which cells create new signaling pathways1,2, but recently duplicated proteins typically must become insulated from each other, and from other paralogs, to prevent unwanted cross-talk3. A similar challenge arises when new sensors or synthetic signaling pathways are engineered within cells or transferred between genomes. How easily new pathways can be introduced into cells depends on the density and distribution of paralogous pathways in the sequence space defined by their specificity-determining residues4,5. Here, we directly probe how crowded sequence space is by generating novel two-component signaling proteins in Escherichia coli using cell sorting coupled to deep-sequencing to analyze large libraries designed based on coevolution patterns. We produce 58 new insulated pathways, in which functional kinase-substrate pairs have different specificities than the parent proteins, and demonstrate that several new pairs are orthogonal to all 27 paralogous pathways in E. coli. Additionally, we readily identify sets of 6 novel kinase-substrate pairs that are mutually orthogonal to each other, significantly increasing the two-component signaling capacity of E. coli. These results indicate that sequence space is not densely occupied. The relative sparsity of paralogs in sequence space suggests that new, insulated pathways can easily arise during evolution or be designed de novo. We demonstrate the latter by engineering a new signaling pathway in E. coli that responds to a plant cytokinin without cross-talk to extant pathways. Our work also demonstrates how coevolution-guided mutagenesis and sequence-space mapping can be used to design large sets of orthogonal protein-protein interactions.
Keywords: protein evolution, protein-protein interactions, signal transduction, synthetic biology, two-component signaling
Many promising new therapies, such as CAR-T cells6 and engineered probiotics7, require an ability to repurpose and transfer signaling pathways into a new genomic context8-10. Similarly, new pathways arise during evolution through the duplication and diversification of existing signaling mechanisms. For engineered and evolved signaling proteins to execute independent functions within cells, they must minimize detrimental cross-talk, a significant challenge for proteins with paralogs.
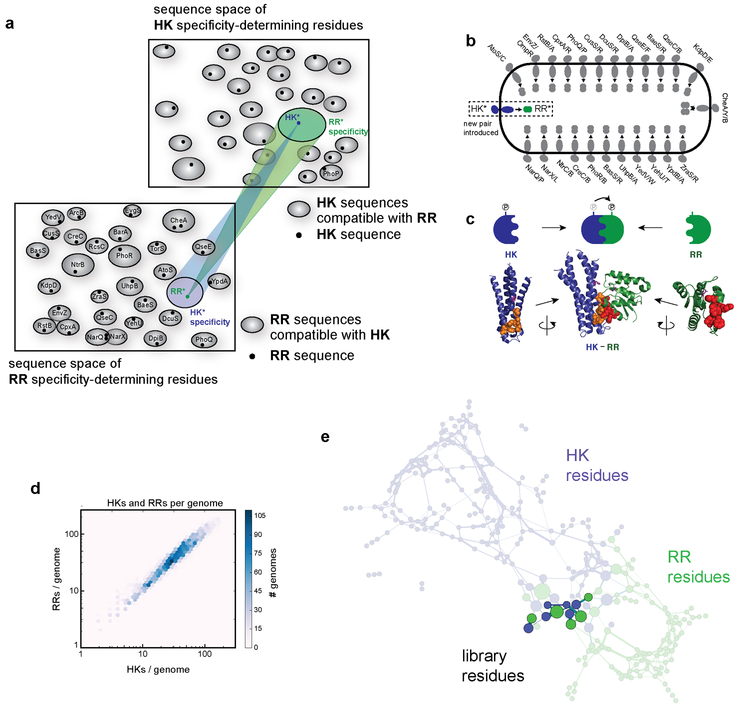
For signaling proteins, such as protein kinases and their substrates, specificity is enforced primarily at the amino-acid level11,12. These specificity-determining residues define a finite sequence space. How cells globally organize paralogous protein families in sequence space and whether the specificity-determining residues of individual members have been optimally distributed to minimize cross-talk during evolution remain open questions. Work with SH3 and PDZ domains in eukaryotes suggested that paralogs are densely packed in sequence space, with intense negative selection against cross-talk leading to a global optimization of specificity4,5. However, sequence space is vast and nature may not have fully occupied or explored it.
To assess how crowded paralogs are in sequence space, we sought to engineer protein complexes that are functional, yet insulated from extant paralogs. If sequence space is densely occupied by existing paralogs, it should be difficult to introduce new, insulated pathways (Fig. 1a, Extended Data 1a). However, if sequence space is sparsely occupied, new pathways should be easy to introduce, with a low probability of cross-talk. The design of orthogonal interacting proteins remains a major challenge with prior efforts have only generated 3-4 orthogonal pairs13-15.
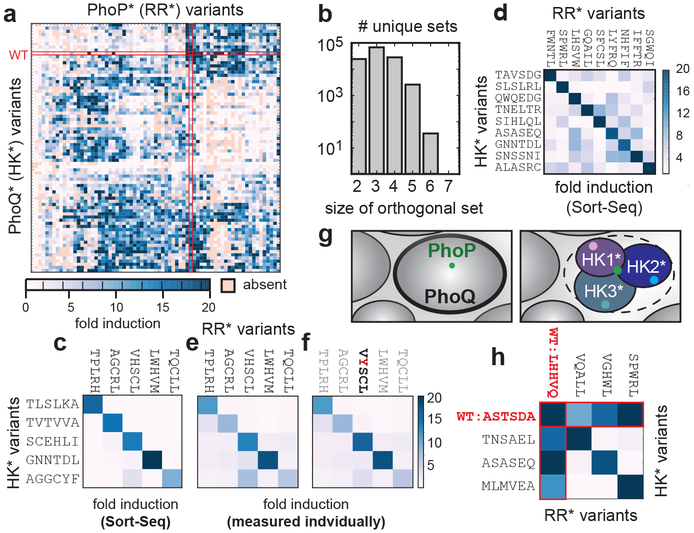
Fig. 1. Probing the density of paralogs in sequence space by building new, orthogonal signaling protein pairs.
(a) Two models for the distribution of paralogs in sequence space. Each oval is a niche representing the set of substrates that a kinase can interact with given its specificity-determining residues. These niches could be densely packed in sequence space (top) or more sparsely distributed (bottom), making the introduction of new, insulated kinases (HK*) relatively difficult or easy, respectively. (b) E. coli PhoQ (blue) can phosphorylate or dephosphorylate PhoP (green), depending on Mg2+ levels, to stimulate or repress, respectively, gene expression, including a YFP reporter (bottom). (c) A library of ~5×108 PhoQ-PhoP variants was first examined by flow cytometry before growth overnight in low Mg2+, followed by outgrowth with high or low Mg2+ and then FACS, with deep sequencing of 8 consecutive bins. For each variant pair, the number of reads in each bin was plotted to infer its fold-induction value. (d) Pearson correlation between YFP values inferred by Sort-seq and measured individually. Points indicate mean±s.d., n=2 biological replicates. (e) For each variant pair indicated, including the wild-type pair (top), the signaling profile inferred by Sort-seq (left) is compared to that measured in isolation (repeated three times with similar results). (f) Plot of mean YFP level inferred by Sort-seq for each variant pair in the OFF and ON states.
We focused on bacterial two-component signaling proteins, which involve a sensor histidine kinase that, upon activation, autophosphorylates and then transfers its phosphoryl group to a cognate response regulator to effect changes in cellular behavior (Fig. 1b, Extended Data 1b-c)16. Most histidine kinases are bifunctional, acting as phosphatases in the absence of a signal to dephosphorylate their cognate regulator. Bacteria usually encode dozens of two-component pathways (Fig. 1a, Extended Data Fig.1d) that are mutually insulated, with the vast majority of kinase-regulator pairs forming exclusive one-to-one relationships2. Both kinase and phosphatase activities, which involve the same protein-protein interface, contribute to pathway specificity11,17,18.
The specificity of kinase-regulator interactions is driven by a limited set of interfacial residues in each protein that strongly coevolve (Extended Data Fig. 1c)19-21. To identify combinations of these residues that are functional and insulated from existing pathways in E. coli, we constructed a dual library of mutants in which the 11 key, coevolving interface residues of a canonical two-component system, PhoQ-PhoP, were randomized (Fig. 1c, Extended Data Fig. 1c-e)20,22.
To identify functional combinations of residues, we first grew the library of PhoQ-PhoP variants overnight in medium with low Mg2+, which activates PhoQ. Because cells must phosphorylate PhoP to grow when Mg2+ is limiting, this step enriches for functional PhoQ-PhoP variants (Fig. 1c). Variants that survived limiting Mg2+ were then subjected to Sort-seq, using fluorescence-activated-cell-sorting and deep-sequencing to quantify their signal responsiveness (Extended Data Fig. 2-3). To gauge PhoP activity in vivo, we used a fluorescent reporter, PmgrB-yfp (Table S1, Table S2). In low extracellular Mg2+, functional PhoQ promotes PhoP phosphorylation and production of YFP, whereas with high Mg2+, PhoQ drives PhoP dephosphorylation, limiting YFP accumulation (Fig. 1b).
To identify those variants that are signal responsive, we sorted cells from each condition into 8 separate bins and deep sequenced the randomized regions of variants in each bin (Fig. 1c). We calculated the frequency of each variant in each bin to yield distributions of individual variants in low and high Mg2+. We then assessed mean YFP levels in each condition and the fold-induction, or signal responsiveness, of each variant.
To validate our selection scheme, we isolated at random 48 individual clones from our starvation-enriched library and measured the distribution of YFP levels in low and high Mg2+ (Fig. 1d). Of these 48 clones, 32 had sufficient sequencing coverage for quantification by Sort-seq. Each individual flow cytometry profile showed strong similarity to the distribution inferred from Sort-seq, including for variants with high fold-induction values and those that were constitutively ON (Fig. 1e). Constitutively ON behavior likely arises when a PhoQ variant lacks phosphatase activity as PhoP can then accumulate phosphoryl groups from other sources, e.g. acetyl-phosphate; such behavior is seen with a ∆phoQ strain21 or variants harboring a stop codon in phoQ (Fig. 1f).
To select variants that are signal responsive like wild-type PhoQ-PhoP, we identified combinations of residues that produced fold-induction values >20; there were 502 such sequences, hereafter called functional PhoQ*-PhoP* variants (Fig. 1f, Extended Data 2i). Most residue combinations identified as functional shared few identities with wild-type PhoQ-PhoP. We isolated and characterized 41 diverse PhoQ*-PhoP* variants that showed >20-fold induction and shared fewer than 5 identities with the wild-type PhoQ-PhoP at the 11 randomized positions (Fig. 2a). To test whether these PhoQ*-PhoP* pairs were insulated from the wild-type proteins, we built strains in which each PhoQ* and PhoP* variant was tested for interaction with a wild-type partner (Table S3). For 16 of the 41 pairs tested, there was substantially higher Mg2+-dependent signaling with the mutant protein pair than for either mutant paired with a wild-type partner (Fig. 2a). Thus, these 16 pairs do not cross-talk with the wild-type PhoQ-PhoP proteins, indicating that our selection scheme produced functional, signaling pathways insulated from the parental proteins.
Fig. 2. Orthogonality of selected PhoQ-PhoP variant pairs.
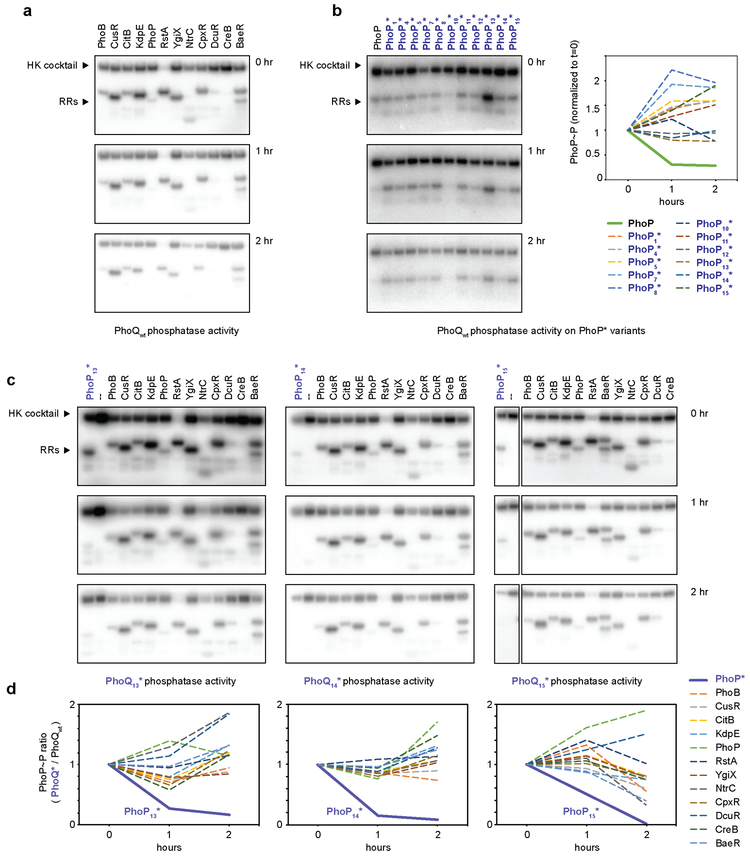
(a) Fold-induction values measured by flow cytometry for PhoQ*-PhoP* pairs (right), compared to PhoQ* with wild-type PhoP (left) and wild-type PhoQ with PhoP* (middle). The interface residues of each pair are listed with the number of identities to the wild-type proteins. (b) Time-course of autophosphorylated PhoQ or PhoQ13* incubated with PhoP or PhoP13*. For PhoQ*-PhoP* and PhoQ-PhoP, phosphotransfer leads initially to phosphorylated PhoP and a concomitant decrease in phosphorylated PhoQ. PhoQ eventually becomes unphosphorylated and drives dephosphorylation of PhoP, yielding a blank lane. (c) PhoQ* variants with no detectable autokinase activity specifically dephosphorylate their cognate PhoP* variants. Phosphorylated PhoP* variants were incubated with their cognate PhoQ* or wild-type PhoQ for the times indicated. (d) Phosphotransfer profiles of wild-type PhoQ (top) and three PhoQ* variants. In each case, autophosphorylated kinase was incubated for 5 min. with each of 27 E. coli response regulators, and its selected PhoP* variant. All gels are representative of >=2 replicates.
To examine the mechanistic basis of insulation, we purified the PhoP* variants and the cytoplasmic domains of the PhoQ* variants for 7 of the 16 validated, insulated protein pairs. The autokinase, phosphotransfer, and phosphatase activities of a histidine kinase can be assessed using a single assay (Fig. 2b)11,16. Each kinase is autophosphorylated with [32P-γ]-ATP and then mixed with a given partner, resulting in phosphotransfer and a phosphorylated response regulator; as unphosphorylated kinase accumulates, it stimulates the dephosphorylation of a response regulator, leading to depletion of radiolabeled response regulator.
For three of the functional mutant pairs, we observed the same pattern of activities as with wild-type proteins, demonstrating that these PhoQ*-PhoP* pathways harbor functional phosphotransfer and phosphatase activities (Fig. 2b, Extended Data 4a). We then tested each mutant protein with a wild-type partner. We found that the wild-type PhoQ could phosphotransfer to some PhoP* variants, but could not efficiently dephosphorylate them. Similarly, we found that the PhoQ* variants could phosphorylate wild-type PhoP, but could not dephosphorylate it. Thus, the orthogonality of these variant pairs with respect to the wild-type system is driven largely by highly-specific phosphatase activity.
For four of the functional, insulated mutant pairs purified, the kinases exhibited no detectable autokinase activity in vitro. However, they retained phosphatase activity and each was specific for the selected, cognate PhoP* partner relative to the wild-type PhoP (Fig. 2c, Extended Data 4b). Thus, the phosphatase activity of a PhoQ* variant may be sufficient to support a functional and insulated PhoQ*-PhoP* system; phosphoryl donors such as acetyl-phosphate (or other kinases) would drive PhoP* phosphorylation in low Mg2+, with PhoQ* phosphatase activity ensuring that PhoP* is not active in high Mg2+.
Next, we assessed the orthogonality of functional variants with respect to the other ~30 two-component signaling pathways in E. coli. We purified 27 response regulators from E. coli and assayed, in parallel, the ability of PhoQ13* and PhoQ15* to phosphorylate each regulator and their partners, PhoP13* and PhoP15*, respectively (Fig. 2d). Strikingly, no phosphotransfer was detected from either kinase to any of the endogenous E. coli response regulators after 5 minutes except the wild-type PhoP. However, as noted above, the PhoQP13* and PhoQP15* pairs are insulated from the wild-type PhoQP by the highly-specific phosphatase activity of each kinase. Some phosphotransfer to other regulators was detected after longer incubations, when many histidine kinases exhibit promiscuity11, reflecting their homology (Extended Data Fig. 4c). We also examined phosphotransfer from 5 E. coli kinases to 11 different PhoP* variants (Extended Data Fig. 5); in each case, the native kinase preferentially phosphorylated its cognate response regulator. PhoP* variants were typically more insulated than wild-type PhoP from these noncognate kinases. Finally, we phosphorylated 12 E. coli response regulators and then examined their dephosphorylation by three PhoQ* variants (Extended Data Fig. 6); in each case, PhoQ* robustly dephosphorylated its cognate PhoP*, but not the native response regulators. Taken together, these results indicate that the functional PhoQ*-PhoP* variants identified are insulated from native E. coli pathways.
To further test the global insulation of selected PhoQ*-PhoP* variants, we used RNA-Seq to examine gene expression when strains carrying one of six different PhoQ*-PhoP* variant pairs were grown with excess or limiting Mg2+ to repress or stimulate PhoQ, respectively. Wild-type and PhoQ*-PhoP* systems produced similar induction of known PhoP-dependent genes (Extended Data Fig. 7a). To assess whether a variant PhoQ* cross-phosphorylated other response regulators, we took advantage of the fact that active response regulators typically autoregulate, promoting expression of themselves and their cognate histidine kinase2. Notably, none of the six strains tested showed significant induction of other two-component systems relative to a wild-type control (Extended Data Fig. 7b-d). These RNA-Seq analyses further indicate that the PhoQ*-PhoP* systems identified are globally insulated from other pathways. Thus, there are unoccupied regions of sequence space where new systems with novel interaction specificities can be introduced without producing cross-talk to existing systems.
We also wanted to test the insulation of our selected, functional PhoQ*-PhoP* variants with respect to each other. To this end, we selected 79 variant pairs that had high fold-induction values and broad sequence diversity. We then combinatorially combined these PhoQ* and PhoP* variants, producing a library with a theoretical diversity of 5,609. This library was transformed into cells harboring the PmgrB-yfp reporter and subjected to Sort-seq as before (Fig. 1c, Extended Data 8a-b), allowing us to infer the fold-induction of each PhoQ*-PhoP* combination. Within the resulting interaction matrix (Fig. 3a, Table S4), 58 variant pairs were orthogonal to the wild-type proteins (Extended Data Fig. 8c).
Fig. 3. Identification of sets of orthogonal signaling proteins.
(a) Fold-induction values for all possible pairings from a set of 79 PhoQ*-PhoP* variants. The matrix was clustered in both dimensions. (b) Number of unique sets of various sizes of orthogonal PhoQ*-PhoP* pairs, with fold-induction values >15 for cognate pairs and <6 for all non-cognate pairs. (c) A set of 5 PhoQ* and PhoP* variants that are functional and mutually orthogonal. (d) A set of 9 PhoQ* and PhoP* variants that are functional and mutually orthogonal with fold-induction values >17 for cognate pairs and <10 for non-cognate pairs. (e) Fold-induction values for the mutant combinations in (c) measured individually. (f) Same as (e) but with a point mutant, PhoP* VHSCL to VYSCL, that reduces cross-talk. (g) Model for subfunctionalization of PhoQ in sequence space. (h) Subfunctionalization of PhoQ specificity. A set of three PhoQ*-PhoP* variants that are mutually insulated, but retain interactions with the parent proteins.
To isolate orthogonal sets of PhoQ*-PhoP* variants, we searched the 79×79 interaction matrix for sub-matrices in which strong interactions were seen only along the diagonal. We isolated more than 2,500 unique sets of 5 orthogonal signaling pairs and dozens of sets of size 6 (Fig. 3b-c, Extended Data Fig. 8d-e). With a slightly relaxed threshold for non-cognate interactions, we found sets with up to 9 orthogonal protein pairs (Fig. 3d). To verify the orthogonality of these sets of PhoQ*-PhoP* variants, we cloned and analyzed the 25 individual pairs comprising a specific 5×5 matrix. Flow cytometry analysis showed strong agreement with the fold-induction values inferred by Sort-seq (Fig. 3c,e).
Notably, the non-cognate pairs in Fig. 3 were measured in the absence of each variant’s cognate partner. Any weak cross-talk seen should be eliminated by the phosphatase activity of the cognate kinase17,18, if present; this prediction was confirmed for one instance of a PhoP* variant that exhibited modest cross-talk from wild-type PhoQ unless its cognate PhoQ* was also expressed (Extended Data Fig. 8f-g). Thus, the off-diagonal values seen with orthogonal sets in Fig. 3b-e represent upper limits on cross-talk. This small degree of cross-talk is also easily reduced further. For example, with the set in Fig. 3e, we screened point mutations of PhoP*(VHSCL) for reduced cross-talk, finding that PhoP*(VYSCL) had reduced interaction with non-cognate kinases while maintaining interaction with its cognate kinase PhoQ*(SCEHLI) (Fig. 3f).
The 79×79 matrix of interactions (Fig. 3a) also offered insight into how new pathways can arise through sub-functionalization, the partitioning of a niche in sequence space rather than movement into a new region (Fig. 3g). For example, we found three pairs of PhoQ*-PhoP* variants that were insulated from each other, but with each exhibiting substantial cross-talk to the parental, wild-type proteins (Fig. 3h, Extended Data Fig. 8h-k). Thus, these pairs have effectively partitioned the original niche of wild-type PhoQ (and PhoP) in sequence space, yielding three insulated pathways. Sub-functionalization of duplicated proteins derived from a promiscuous ancestor may be a common mechanism by which insulated paralogs arise during evolution.
Collectively, our results indicate that the sequence space of two-component signaling pathways in E. coli is relatively sparsely occupied, such that new, orthogonal signaling pathways can readily be introduced. The 502 functional variant pairs isolated here have few specificity residues in common with wild-type PhoQ-PhoP, each other, or extant two-component signaling proteins (Extended Data Fig. 9a). A force-directed graph based on specificity residue similarity highlights the diversity of naturally-occurring interfaces and the variants we isolated (Extended Data Fig. 9b). To estimate how easily a new, insulated pathway can be introduced, we noted that 502 functional pairs came from 10,595 pairs with quantifiable fold-induction values. Of these 502, ~40% are likely insulated from wild-type PhoQ-PhoP (Fig. 2a), implying that ~200 (1.6%) of the 10,595 are both functional and insulated. This frequency is an upper-bound as the initial 10,595 pairs arose from low Mg2+ selection (Fig. 1c), which enriches ~100-fold for functionality (Extended Data Fig. 2b). Nevertheless, given the size of sequence space, these estimates underscore the relative ease of creating kinase-substrate pairs that are functional and orthogonal to their parent proteins.
Orthogonal signaling pathways will be useful in generating synthetic sensors and novel regulatory systems. As an example, we sought to generate a new pathway in E. coli that responds to the cytokinin trans-zeatin, a plant hormone. The histidine kinase AHK4 from Arabidopsis thaliana senses trans-zeatin, but cross-talks extensively with a native two-component pathway in E. coli23. To overcome this limitation, we fused the AHK4 sensory domain to the kinase domains of an orthogonal PhoQ* and expressed this construct in E. coli with the cognate PhoP* (Fig. 4a-b). This engineered sensor kinase enabled E. coli to respond specifically to trans-zeatin and it was insulated from all native two-component pathways, as measured by phosphotransfer profiling and RNA-seq (Extended Data Fig. 10). Thus, this chimeric sensor kinase and its cognate PhoP* expand the sensory repertoire of E. coli without introducing undesirable cross-talk.
Fig. 4. Construction of an insulated sensor.
(a) An insulated cytokinin (trans-zeatin) sensor constructed by fusing the sensory domain of Arabidopsis thaliana AHK4 to PhoQ4*. (b) Chimeric sensor AQ4* is specifically responsive to 1 μM trans-zeatin, phosphorylating its cognate mutant PhoP4* to activate a YFP reporter. Wild-type PhoQ responds only to Mg2+ and not to the cytokinin. Bars indicate mean from n=3 biological replicates.
Synthetic circuits have, to date, been built mainly from nucleic-acid components because of their intrinsic modularity and programmability24. Protein-based circuits offer faster response times and richer functionality, but require more complicated programming of protein interactions. Our work now enables the design of two-component signaling-based circuits in bacteria or eukaryotes. And the relatively sparse distribution of paralogs in sequence space means multiple pathways can readily be introduced.
In sum, our work highlights the power of using coevolution-guided libraries to investigate protein-protein interactions and supports a model in which sequence space is not densely occupied. The relatively sparse distribution of extant proteins in sequence space presumably reflects their evolutionary history. A prior study indicated that duplicated signaling proteins are under pressure immediately post-duplication to change and become insulated, but subsequent movement in sequence space then arises only from neutral changes3. Although duplicated proteins are initially subject to selection against cross-talk with each other, each protein is likely not subject to system-wide negative selection or global optimization.
Methods
Bacterial strains and media
Escherichia coli strains were grown in M9 media (1x M9 salts, 100 μM CaCl2, 0.2% glucose, 0.1% casamino acids and MgSO4 at indicated concentrations). When indicated, antibiotics were used at the following concentrations: carbenicillin, 50 μg/mL; kanamycin, 50 μg/mL; spectinomycin, 50 μg/mL, chloramphenicol, 32 μg/mL.
The base strain for all studies was E. coli strain TIM171 (MG1655 ΔphoPQΔlacZYA attλ::[PmgrB-yfp] attHK::[PtetA-cfp+]) with a ColE1/ampR plasmid (pCM150) containing PmgrB-yfp. All libraries were cloned onto a low copy pSC101/specR plasmid (pCM099, a derivative of pLPQ2), where phoPQ was driven by a constitutive lacUV5 promoter. We also introduced a bicistron RBS (BCD18) upstream of phoPQ25, which leads to expression of a single transcript encoding a small (17 a.a.) ORF followed by an independent ribosome binding site and then phoPQ. This configuration ensures that mutations near the 5’ end of the phoP coding region do not substantially affect expression by changing interactions between the 5’ end of phoP and the upstream leader sequence. Expression from the lacUV5 promoter on a plasmid likely produces more PhoQ-PhoP than natively produced, increasing the chance of cross-talk; thus, the variant pairs identified as orthogonal would perform even better with respect to cross-talk at lower expression levels. Additional characterizations of PhoQ* and PhoP* variants isolated from the library were done with a three-plasmid setup: reporter plasmid pCM150, pCM143 (lacUV5-BCD18-phoP, pSC101/specR), and pCM149 (lacUV5-RBS_B0034-phoQ, p15A/kanR). Point mutations were introduced using blunt end ligation26 and Gibson assembly27.
Flow cytometry characterization
To induce PhoPQ, cells were grown to mid-exponential phase (OD600 ~ 0.5) in M9 before being washed once with M9 containing 0 mM MgSO4 and diluted 1:100 into M9 containing 10 μM MgSO4 (for ON/induction) or 50 mM MgSO4 (for OFF/repression). Cells were grown for 6 hr, diluted 1:50 into PBS with 0.5 g/L kanamycin, and fluorescence measured on a Miltenyi MACSQuant VYB. An identical procedure was used to induce AHK4-PhoQ fusions, except cells were grown in M9 containing 2 mM MgSO4 and 1 nM aTc (anhydrotetracycline, Sigma) at all times, with the ON condition containing 1 μM trans-zeatin (Sigma) and the OFF condition containing no trans-zeatin. In each cytometry experiment, three replicates of each sample were induced independently and 20,000 cells were measured per replicate. FlowJo was used to analyze the data, gating on single, live cells and extracting the geometric mean of the YFP distribution (Extended Data Fig. 2j). Error bars indicate the standard deviation of the geometric means measured in each replicate.
Design and assembly of degenerate PhoQ-PhoP library
The PhoQ-PhoP saturation mutation library was constructed by replacing the targeted residues with NNS codons28-30. The residues targeted were selected, as indicated in the main text, based on amino acid coevolution analyses performed using GREMLIN31.
The plasmid library was assembled in two general steps: 1) individual PhoP and PhoQ libraries were built in separate vectors and 2) sections of these vectors were combined to produce a new vector containing both PhoP and PhoQ mutants (Extended Data Fig. 2a). For the first step, oligonucleotide libraries for the sections of phoP and phoQ to be mutated were ordered from DNA 2.0. NNS nucleotides replaced codons 12, 14, 15, 18, and 19 in PhoP and codons 284, 288, 289, 292, 302, and 303 in PhoQ. These oligonucleotides were cloned into vectors pCM071 and pCM076 using the Type IIS restriction enzyme BsmBI. A toxic ccdB locus on these plasmids, used as a counter selection, was replaced during the process, ensuring a high rate of insertion incorporation. Both insert and vector were digested with BsmBI at 55°C for 2 hr and then purified on a Zymo DNA clean column. 1 pmol of both insert and vector were combined in a 25 μL reaction with 400 units of T4 ligase and incubated at 16 °C for 16 hr. Three ligations of each library were done, to ensure sufficient numbers of transformants. Ligations were dialyzed on Millipore VSWP 0.025 μm membrane filters for 60 min. and then the entire volume was electroporated into 20 μL of Invitrogen MegaX DH10B cells. From three ligations of each library, a total of 2.3 × 108 and 7.4 × 107 transformants were obtained for the PhoQ and PhoP libraries, respectively.
BsaI sites were used to join the two sublibraries into a single plasmid. The fusion points were designed such that faulty assemblies would not be viable: one junction was within the specR cassette (both the PhoP and PhoQ sublibraries harbored a kanR cassette, but only half the specR open reading frame) and the other was within PhoQ. 500 fmol of minipreped DNA product (Qiagen) from each of the two libraries were combined in 25 μL T4 ligase buffer and digested with 1 μL BsaI for 1 hr at 37 °C. T4 ligase was then added and the reaction was cycled between 16 °C (3 min) and 37 °C (2 min) for 50 cycles to allow iterative ligation and digestion, running the reaction to completion. Final ligation product was dialyzed on Millipore VSWP 0.025 μm membrane filters for 60 min and the entire volume electroporated into 20 μL of Invitrogen MegaX DH10B cells. In total, 12 ligations and electroporations were done to produce a total of 5.72 × 108 transformants. Transformations were pooled and grown overnight (14 hr) in 100 mL 2xYT + carbenicillin and spectinomycin. Following assembly, the plasmid library was purified by miniprep (Qiagen), dialyzed, and electroporated into ∆phoPQ strain CJM2044 to yield 3.8 × 109 transformants.
Library selection and Sort-seq
The PhoQ-PhoP library was subjected to an initial selection of Mg2+ starvation to enrich for functional variants before performing fluorescence activated cell sorting (FACS). To this end, 6 mL of an overnight culture of the library (in 2xYT) was washed in M9 and diluted to an OD600 ~ 0.1 in 100 mL M9 containing 2 mM MgSO4. Three replicates of this culture were made, and carried separately through the subsequent selection, FACS, and deep sequencing. Cells were grown for approximately 2 hours to OD600 ~ 0.4. at which point 1.6 mL of culture was washed three times in M9 containing no MgSO4, and used to inoculate 100 mL of M9 containing no MgSO4. After each dilution, the culture was sampled and a dilution series was plated on LB plates to ensure no bottlenecking occurred (CFUs > 1 × 109). The cultures in M9 containing no MgSO4 were grown overnight (14 hr), with the OD600 increasing from 0.05 to only ~0.07. MgSO4 was then added to bring the concentration to 2 mM, and cells were grown to an OD600 of 0.5 in 6 hrs, at which point glycerol stocks were made.
For FACS, 1 mL glycerol stocks were thawed and inoculated into 25 mL of M9. For each library replicate, one frozen stock aliquot was added directly to M9 containing 50 mM MgSO4 (OFF state) and one aliquot was washed three times in M9 containing 0 mM MgSO4 before inoculation into M9 with 10 μM MgSO4 (ON state). To maintain cells in exponential phase, cultures were diluted (1:4 for ON state, 1:10 for OFF state) after 3 hours. After 6 hrs, cells were diluted again (1:5), and chloramphenicol was added to a concentration of 320 μg/mL and cells were placed on ice for sorting. CFP was expressed at a low constitutive level (attHK::[PtetA-cfp]), and used to normalize YFP expression. Cells were sorted into ratiometric bins on the diagonal of CFP and YFP expression, to control for extrinsic expression noise in the YFP signal. For each library replicate, both the ON and OFF cultures were sorted into 8 separate bins, generating 48 total bins. Up to 2.5 million cells were sorted into bins per replicate (Extended Data Fig. 2c, 8a). Sorted cells were added to 2xYT media containing 2 mM MgSO4, carbenicillin, and spectinomycin, and then grown overnight.
Illumina sample preparation
After FACS, plasmids were purified (Qiagen MiniPrep) from overnight cultures representing each bin from each library replicate. For the two mutagenized regions of the plasmid to be brought into close enough proximity (< 790 bp) for paired-end Illumina sequencing (Extended Data Fig. 2b), plasmids were digested with XhoI and then self-ligated (T4 ligase, 4 hr). To isolate only self-ligation products, and not cross-ligation products, ligation reactions were cleaned (Zymo PCR Clean Up) and gel purified to select for the correct size on FlashGels (Lonza). Two PCR reactions were performed, both using KAPA HiFi Hotstart, to add Illumina sequencing adaptors and barcodes. First, ligation reaction products were amplified for 30 cycles (95 °C for 30 s, 65 °C for 15 s, 72 °C for 120 s) with primers CJM642 and CJM643 in an emulsion PCR (Micellula Emulsion PCR) to avoid PCR chimeras. Second, purified PCR product from the first reaction was subjected to a second PCR with barcoding primers for 9 cycles (95 °C for 30 s, 65 °C for 15 s, 72 °C for 60 s). Final products were quantified (NanoDrop), normalized, combined, and sequenced on an Illumina NextSeq. For each bin, 1-33 million reads were collected.
Construction of combinatorial mutant library
79 pairs of PhoQ*-PhoP* variants were selected that displayed broad sequence diversity and high fold induction in the initial PhoQ*-PhoP* library. These pairs included 79 unique PhoQ* variants and 71 unique PhoP* variants. These unique variants were cloned into plasmids pCM143 (PhoP) and pCM149 (PhoQ) using blunt end ligation26 and Gibson assembly27. Each pCM143 and pCM149 variant was amplified by PCR (KAPA Hifi, 30 cycles) to generate amplicons for Gibson assembly (primers CM937/CM1531 for pCM149 and primers CM938/CM1532 for pCM143). PCR products were cleaned (Zymo PCR Cleanup), quantified by NanoDrop, and combined into an equimolar mix of pCM143 amplicons and an equimolar mix of pCM149 amplicons. The two mixes were combined in Gibson Assembly master mix (300 fmol of large pCM143 fragment, 900 fmol of smaller pCM149 insert), incubated at 50 °C for 2 hr and heat killed at 79 °C for 20 min. The assembly was dialyzed on Millipore VSWP 0.025 μm membrane filters for 60 min and transformed into electrocompetent CJM2044 cells.
Unlike the treatment of the initial, larger library, this library was not subjected to a low magnesium selection step. Immediately after construction, this library underwent Sort-seq, as described above.
Illumina data processing
The frequency of each mutant in each bin was calculated by taking the fraction of reads in a given bin corresponding to a given sequence, normalized by the fraction of cells in that given bin (see Supplementary Information). All Sort-seq plots display the mean frequencies in each bin across three replicates, with error bars indicating standard deviation. Gaussian functions were fit to each distribution (in log10YFP units), from both the ON and OFF sorts (SciPy optimize package). Variants with fewer than 25 total reads were discarded before fitting. Poor Gaussian fits have high variances on the estimated parameters. The standard deviation error on the estimated log(YFP) mean (σfit) was used as a metric to filter poorly fit sequences: sequences were removed if σfit,ON + σfit,OFF > 2. In total, 10,595 unique variants passed these filters. Fold-induction values were calculated as the ratio of the fit means between the induced and uninduced states: μON / μOFF.
During analysis of the second, combinatorial library (Fig. 3a), the fold induction was calculated for the most frequent nucleotide sequence representing each amino-acid sequence. For visualization and orthogonal set design, fold inductions were bounded between 1 and 20. Individually tested mutants generally did not surpass the sensitivity of wild type PhoQ-PhoP, which displayed 20-fold induction during Sort-seq, suggesting that signal above 20-fold may be due to noise. The axes of the matrix in Fig. 3a were clustered hierarchically using the WGPMC method (Scipy). During clustering, matrix entries which lacked data were assigned the mean value of all other entries.
Analysis of the sensitivity of Sort-seq quantification to read count
To assess the quality of Sort-seq-based quantification for variants with lower read coverage, variants with high read coverage (2,000-10,000 total reads) were down-sampled to simulate low read coverage and fed through the Sort-seq analysis pipeline (Extended Data Fig. 3). For each of these high-coverage variants, simulated data was produced by down sampling 100 independent times by the factors indicated in Extended Data Fig. 3. Simulated read coverage was generated by sampling (with replacement) from the original reads of each variant up to the desired read coverage. Simulated reads were then subjected to the same Gaussian fitting protocol as before. As in the original analysis, poor fits (σfit,ON + σfit,OFF > 2) were discarded. All simulated variants were classified as functional or non-functional, based on the fold-induction values of original, high coverage variants they were sampled from (Extended Data Fig. 3c, e). False positive rates (Extended Data Fig. 3d) and false negative rates (Extended Data Fig. 3f) were then calculated as a function of read coverage by computing the fraction of simulated variants that were mis-classified.
Orthogonal set design
Orthogonal sets of PhoQ* and PhoP* variants up to 7 pairs were identified by systematically scanning all PhoQ*-PhoP* permutations within the matrix in Fig. 3a and Table S4. Larger orthogonal sets were identified using a greedy search algorithm (see Supplementary Information). The 5×5 orthogonal set described in Fig. 4e was further optimized by testing single point mutants of the single PhoP* variant (VHSCL) that initially displayed cross-talk with a non-cognate PhoQ* variant. Each of the 5 PhoP* specificity residues was replaced independently with an NNK codon to generate all possible single point mutants (xHSCL, VxSCL, VHxCL, VHSxL, VHSCx where × is any amino acid specified by the NNK codon). These mutants were cotransformed with the cognate PhoQ* variant (SCEHLI) into CJM2044 and grown overnight in M9 media containing 0 mM MgSO4 to remove non-functional variants. After plating the surviving strains on LB agarose plates, 48 clones were tested for Mg2+ induction (see below). The pCM143-PhoP* plasmids from the 24 clones with the strongest induction were purified and cotransformed with the non-cognate PhoQ* variant (AGGCYF) into CJM2044. Mg2+ induction was measured by cytometry and the 8 clones displaying the highest cognate / non-cognate induction ratio were selected for testing with all five PhoQ* variants. Two of these eight clones were PhoP* (VYSCL), which displayed the highest specificity.
Reconstruction and in vivo characterization of individual PhoQ* and PhoP* variants
Variants were cloned into plasmids pCM143 (PhoP) and pCM149 (PhoQ) using blunt end ligation26 and Gibson assembly27. Combinations of pCM143 and pCM149 plasmids were co-transformed into strain CM2044. Colonies were grown overnight in M9 containing 2 mM MgSO4 and induced, as described above, with M9 containing either 10 μM or 50 mM MgSO4. After 6 hours, cultures were diluted 1:50 into cold PBS containing 0.5 g/L kanamycin, and fluorescence measured on a Miltenyi MACSQuant VYB. Note that the fold-induction values of individually tested variant pairs were generally smaller than those measured by Sort-seq, likely due to differences between the Miltenyi cytometer and BD Aria Sorter; however, the two measurements were highly correlated (Pearson R2 = 0.91, Fig. 1d).
Purification of two-component signaling proteins and in vitro phosphotransfer assays
Expression and purification of PhoQ* and PhoP* variants, and phosphotransfer experiments were carried out as previously described11,21. PhoP* was purified fused to a His6-Trx tag, and the cytoplasmic region of PhoQ* (residues 238-486) was fused to a His6-MBP (maltose binding protein) tag. For phosphotransfer reactions, the kinase was autophosphorylated for 1 hr at 30 °C with [γ−32P] ATP (Perkin Elmer) before being combined with PhoP* at a 1:8 ratio (10 μL reactions contained 1 μM PhoQ* and 8 μM PhoP*). Reactions were stopped at appropriate times by adding 4x Laemmli buffer with 8% 2-mercaptoethanol. This process allowed monitoring of both phosphotransfer and phosphatase activities between PhoQ* and PhoP* variants (Fig. 2b).
For PhoQ* variants where in vitro autophosphorylation was not observed, phosphatase activity was assayed by mixing a given PhoQ* variant with a PhoP* variant that was phosphorylated using wild-type PhoQ. This was achieved by incubating 8 μM PhoP* for 1 hr at 30 °C with [γ−32P] ATP and 1 μM wild-type PhoQ, which promiscuously phosphorylates, but does not dephosphorylate, most PhoP variants. After generating phosphorylated PhoP*, 1 μM of the PhoQ* variant was added and samples taken and reactions stopped as before.
Phosphatase activity of PhoQ* variants with respect to other response regulators was measured with a similar assay. Twelve E. coli response regulators were selected for their ability to be stably phosphorylated in vitro by a cocktail of six E. coli histidine kinases (CreC, RstA, PhoR, PhoP, EnvZ and CpxA, each at 250 nM). After 2 hours of pre-incubation with radiolabeled ATP and this kinase cocktail, each regulator was combined with 2 μM PhoQ or PhoQ* variant. Reactions were stopped at 0, 60 and 120 minutes by adding 4x Laemmli buffer with 8% 2-mercaptoethanol.
Response regulators for phosphotransfer profiles were purified as described above. Each was fused to an N-terminal His6-Trx tag, expressed in BL21(DE3) cells and purified on a Ni2+-NTA column3. Conditions for phosphophotransfer profiles were also identical to above conditions; 10 μL reactions containing 1 μM [γ−32P] autophosphorylated PhoQ* and 8 μM response regulator were generated and stopped after 5 or 60 minutes. Gel images were analyzed using quantified with ImageJ.
RNA-seq
Cultures were grown overnight in M9 media containing 2 mM MgSO4 and then diluted 1:25 into fresh M9 containing 2 mM MgSO4 and grown for 2 hours to reach OD600 ~ 0.5. For the OFF condition, 1 mL of cells was diluted into 2 mL M9 containing 74 mM MgSO4 for a final concentration of 50 mM MgSO4. For the ON condition, 2 mL of cells were pelleted, washed twice with M9 + 10 μM MgSO4, resuspended in M9 + 10 μM MgSO4, and then 1 mL of cells was diluted into 2 mL M9 + 10μM MgSO4. Induction of AQ4 strains was identical, except cells were induced for 30 minutes in M9 (2 mM MgSO4) containing either 0 μM trans zeatin (OFF condition) or 1 μM trans zeatin (ON condition).
RNA was harvested as previously described32. After 30 minutes of growth, cells from each condition were harvested by adding 1.8 mL culture to 200 μM cold stop solution (95% ethanol, 5% acid buffered phenol, 4 °C). The mixture was centrifuged for 30 s at 13,000 rpm on a benchtop centrifuge, and the supernatant was removed with the pellet flash frozen in liquid nitrogen and stored at −80 °C. To extract RNA, Trizol (Invitrogen) was heated to 65 °C, added directly to the pellet, and incubated at 65 °C for 10 minutes with shaking at 2000 rpm (Eppendorf Thermomixer). The mixture was frozen at −80 °C for at least 10 minutes. After thawing, cells were centrifuged at 15,000 rpm, 4 °C for 5 minutes, and the supernatant was removed into 400 μL ethanol. The mixture was applied to a DirectZol spin column (Zymo) and centrifuged for 30 s at 13,000 rpm. The columns were washed with DirectZol RNA prewash buffer twice (400 μL) and RNA wash buffer (700 μL) once before eluting in 90 μL DEPC water. 10 μL 10x Turbo DNAse buffer and 2 μL Turbo DNAse (Invitrogen) were added to the eluant. The mixture was digested at 37 °C for 20 minutes, followed by the addition of 2 μL more DNAse and another 20 minute incubation. Total volume was brought to 200 μL with DEPC water and combined with 200 μL acid-phenol:chloroform (IAA, Invitrogen), vortexed and centrifuged for 10 minutes at 21,000 g and 4 °C. The top (aqueous) layer was extracted and ethanol precipitated in 20 μL NaOAc (3M), 2 μL GlycoBlue (Invitrogen) and 600 μL cold ethanol. Precipitation mix was incubated at −80 °C for more than 4 hours before centrifuging for 30 minutes at 21,000 g and 4 °C. The pellet was washed twice with 500 μL cold 70% ethanol, then air dried and resuspended in 50 μL DEPC water. RNA integrity was validated on a 6% TBE-urea acrylamide Novex gel (Invitrogen) and yield was quantified by NanoDrop spectrophotometer. rRNA was removed with the RiboZero rRNA Removal Kit for Bacteria (Illumina). RNA was fragmented and cDNA libraries were prepared at the MIT BioMicro Center sequencing core using the KAPA RNA HyperPrep Kit (Roche) and sequenced on an Illumina HiSeq. Reads were mapped to the E. coli genome and plasmids with bowtie2 using default parameters33.
To determine whether selected PhoQ*-PhoP* variants interfered with other two-component signaling pathways, we examined whether any other response regulator or histidine kinase genes were upregulated transcriptionally when PhoQ* variants were activated by low Mg2+. We calculated the fold change in expression of each two-component regulatory gene as a ratio of reads in low and high Mg2+ (Extended Data Fig. 7b). Note that rstA and rstB are part of the PhoP regulon and are directly upregulated by wild-type PhoQ-PhoP and most variant pairs. To quantify how these fold changes compared to wild-type PhoQ-PhoP, we calculated the ratio the fold change in each gene to the geometric mean of the fold change in the same gene for the two wild-type replicates (Extended Data Fig. 7c). To assess whether any two-component signaling genes were significantly upregulated, we calculated the Z-score of each ratio of fold changes and conducted a one-tailed test to compute p values (Extended Data Fig. 7d). After using a Bonferroni correction for multiple hypothesis testing, no genes were found to be significantly upregulated (p < 0.05).
Identification of two-component signaling proteins and generation of force-directed graphs
The RefSeq Prokaryotic Genomes database of 5,506 bacterial genomes (Sept, 2017) was downloaded from NCBI. The database was scanned for histidine kinases and response regulators using jackhmmer 34 (E-value cutoff = 0.01) with all two-component signaling proteins from E. coli used as queries. The combined lists of HK and RR hits were aligned with hmmalign 34 to the PFAM hidden Markov models for HisKA and Response_Reg domain families, respectively. Columns in the multiple sequence alignment with greater than 80% gaps were eliminated, and sequences with greater than 50% gaps were discarded. Histidine kinases lacking the catalytic histidine and response regulators lacking the catalytic aspartate were removed. Proteins containing both the HK DHp domain and a RR receiver domain were discarded to avoid ambiguity. HKs and RRs were then labeled as exclusive pairs if they were (i) within 20 genes in the genome, with no other HK or RR genes between, (ii) on the same strand, and (iii) closer to no other potential HK or RR partner (with distance defined as the number of genes between partners). The sequences of paired HKs and RRs were concatenated and the multiple sequence alignment was then reduced to the eleven positions mutated in this study.
The force directed graph was generated using the Gephi network visualization package35. To construct a network, the 85,782 co-operonic HK-RR pairs identified by HMMER in bacterial genomes were combined with the functional mutant sequences from the PhoQ-PhoP dual library that had fold-induction values > 18 and the mutant variants within the characterized 5×5 orthogonal set (Fig. 3c, e). These sequences were treated as nodes and were connected by edges if the pairwise alignment score for the two sequences’ 11 specificity residues (using the BLOSUM62 scoring matrix) exceeded a threshold score of 20. If more than 40 edges were connected to a node, only the top scoring 40 edges were kept. If no edge scoring above 20 connected a node, that node retained its top-scoring edge, despite that edge being below the BLOSUM62 threshold. A final model of ~86,000 nodes and 2.5 million edges was loaded into Gephi and visualized using the Force-Atlas-2 tool35.
Construction of AHK4-PhoQ chimera
Chimeric histidine kinases sensors were made using a variation of the PATCHY strategy (primer aided truncation for the creation of hybrid proteins)36. The N-terminal region of AHK4 (residues 1-475) were cloned downstream of the Ptac promoter on a p15A/kanR vector. This plasmid was amplified via PCR with primers containing SapI sites to allow insertion of the PhoQ kinase domain. Five distinct sets of primers allowed five possible junction sites within AHK4 (residues A466, A468, A469, A472 and A478) with identical GCG overhangs. PhoQ (pCM149) was amplified with 32 distinct primers (also containing SapI sites) to generate 32 C-terminal truncations beginning upstream of the DHp domain (residues 213-224, 257-276). PCR products were gel-purified (Zymo), then combined in a 50 μL ligation reaction containing 400 U of T4 ligase (NEB), 20 U Sap1 (NEB), 100 fmol pooled AHK4 PCR products, and 500 fmol pooled PhoQ PCR products. The reaction was cycled 50 times between 37 °C (2 min) and 16 °C (3 min) to drive assembly to completion, heat killed at 50 °C (20 min) and 80 °C (20 min), and dialyzed on Millipore VSWP 0.025 μm membrane filters (60 min). This small library of 160 possible fusions was transformed into electrocompetent CJM2044 cells harboring PhoP on a plasmid (pCM143).
To enrich for chimeras responsive to trans-zeatin we used Mg2+ starvation as a selection. An overnight culture of the library in M9 media was induced for 1 hour (M9 containing 21 nM aTc, 1 μM trans-zeatin), washed three times in M9 containing no MgSO4 and diluted 1:10 into 100 mL M9 containing no MgSO4, 21 nM aTc, 1 μM trans-zeatin. After 4 hours, 500 μL of culture was plated on LB. 96 colonies were picked and screened for trans-zeatin-dependent YFP expression.
GCN4-DHp fusions to test phosphatase buffering against crosstalk in vivo
To generate cytosolic variants of PhoQ locked in a phosphatase state, we followed a previously described strategy37 and fused GCN4 (MKQLEDKVEELLSKNYHLENEVARL) N-terminal to PhoQ’s DHp domain. pCM149 (lacUV5-phoQ, kanR, p15A) was amplified with 24 distinct primers (containing SapI sites) to generate 24 C-terminal truncations beginning upstream of the DHp domain (residues 222-225, 257-276). The N-terminus of PhoQ was replaced by GCN4 in each of these plasmids, removing the transmembrane and sensory domains. Each GCN4 fusion plasmid was transformed with pCM143 (phoP) and pCM150 (PmgrB-yfp) into TIM175 and tested by standard Mg2+ induction (see above) for activity. As expected, some variants displayed constitutive high YFP (presumably locked kinase conformation) or constitutive low YFP (presumably locked phosphatase conformation) and stepwise amino acid insertions displayed a periodicity of these phenotypes (Extended Data Fig. 8f). One of these fusions (fusion-266) displayed even lower constitutive YFP values than PhoQ with mutations in the ATP cap (R434M, R439M, Q442M, pCM180) or ATP pocket (N385L, N389L, K392M, Y393F, pCM179)38.
To test the ability of a cognate phosphatase to suppress crosstalk from a non-cognate kinase, we used a three-plasmid setup: pCM874 (reporter plasmid pCM150 with PlacUV5-PhoP15* inserted), pCM149 (PlacUV5-PhoQwt) and pCM873 or pCM898 (Ptet-GCN4-fusion266-PhoQwt or -PhoQ15*, respectively). Because PhoP* has been moved from a low-copy to medium-copy plasmid, crosstalk between PhoQwt and PhoP* is likely exacerbated by overexpression, as noted by the high level of induction seen in Extended Data Fig. 8g before aTc is added. However, induction of the GCN4-fusion266-PhoQ15* phosphatase effectively eliminates this cross-talk (Extended Data Fig. 8g, right panel). Induction of the non-cognate phosphatase, GCN4-fusion266-PhoQwt*, does not relieve this cross-talk.
Extended Data
Extended Data Fig. 1. Bioinformatic and coevolutionary analysis of two-component signaling systems.
(a) Schematic illustrating the challenge of identifying new HK* - RR* pairs that are orthogonal to all endogenous HKs and RRs. For both HKs and RRs, the specificity-determining residues define a finite sequence space. For each HK, its specificity residues determine the set of RRs that it can interact with. These sets, or niches in sequence space, are depicted as ovals with each cognate RR represented by a black dot (bottom left). A similar representation is shown for each RR and the set of HKs that it can interact with (top right). The two sequence spaces are connected, as depicted with colored cones for a single HK-RR pair. The establishment of a new signaling pathway that is orthogonal to existing systems requires that the two new proteins are compatible with each other, but occupy regions of HK and RR specificity space that are incompatible with all paralogs already present.
(b) Schematic summarizing the endogenous two-component pathways in E. coli that a new, orthogonal pathway must avoid cross-talk to.
(c) Diagram of the DHp domain of a histidine kinase (HK), TM0853 (blue), in complex with its cognate response regulator (RR), RR0468 (green). Residues that dictate specificity and were randomized in our libraries are spacefilled in orange (kinase) and red (substrate).
(d) Plot summarizing the number of histidine kinases and response regulators in bacterial genomes. (e) Visualization of the GREMLIN model representing the coevolutionary dependencies between the residues of cognate histidine kinases and response regulators. Blue nodes indicate PhoQ residues, green nodes indicate PhoP residues, and the darker nodes are the 11 residues randomized in the dual PhoQ-PhoP library. Edge widths indicate the strength of coevolutionary signal, while node size of each residue represents the total coevolutionary signal to residues on the other protein.
Extended Data Fig. 2. Summary statistics for the dual PhoQ-PhoP library.
(a) Schematic summary of library design.
(b) (Left) Histogram of the read counts for the pre-selection PhoQ-PhoP library. The vast majority of reads are unique, indicating that the library size is larger than Illumina sequencing coverage and no variants are over-represented. (Right) Histogram of the read counts for one replicate of the PhoQ-PhoP library after overnight growth in low Mg2+ conditions.
(c) Counts of cells sorted into each bin, for each replicate and growth condition.
(d) The cells sorted into each bin were grown overnight, diluted back to mid-exponential phase, shifted to media with 10 μM Mg2+ and their YFP levels verified by flow cytometry. n = 2 independent biological replicates.
(e) Same as panel (d), but with cells retained in media with 50 mM Mg2+. n = 2 independent biological replicates.
(f) Scatterplots displaying the correlations between bin frequencies of individual variant pairs measured in independent replicates. Only 106 data points are shown for clarity. R2 values indicate the Pearson correlation coefficients, calculated using all data points.
(g) Same as panel (f), but displaying only the 10,595 variants with sufficient sequencing coverage and fit quality (see Methods) to be included in analysis.
(h) Scatterplot displaying the Pearson correlation between YFP fold-induction measured by Sort-seq and that measured individually by flow cytometry for 32 individual variant pairs.
(i) Sequence logos summarizing the amino acid frequencies at each position varied in the pre-selected library (top), set of pairs with >20-fold induction (middle), and all native HK-RR pairs (bottom). The residues found at these positions in wild-type PhoQ and PhoP are listed below.
(j) The FACS gating strategy for isolating single, live cells for quantification of YFP expression.
Extended Data Fig. 3. Sensitivity of Sort-seq pipeline to read coverage.
(a) Histogram of the total read counts for the 10,595 variants with sufficient coverage and fit quality to be included in analysis.
(b) A schematic example of the down-sampling method used to simulate low read coverage using variants with high read coverage.
(c) Fold induction of high coverage, functional PhoQ*-PhoP* variants (fold induction > 20) after simulating lower read coverage via down-sampling and re-fitting. (n = 100 independent downsampling simulations).
(d) A quantification of how read coverage in (c) affects the calculated fold induction of functional variants. The fraction of functional variants that still display high fold induction at lower read coverage is plotted with respect to read coverage.
(e) Same as panel (c), but for non-functional (fold induction < 20) PhoQ*-PhoP* pairs with high read coverage. (n = 100 independent downsampling simulations).
(f) Same as panel (d), but for non-functional variants. The fraction of non-functional variants that still display low fold induction at lower read coverage is plotted with respect to read coverage.
Extended Data Fig. 4. Additional in vitro characterizations of selected PhoQ*-PhoP* pairs.
(a) Phosphotransfer reactions for PhoQ*-PhoP* variants, as well as PhoQ* with wild-type PhoP (middle), and wild-type PhoQ with PhoP* (right). These experiments were repeated independently two times with similar results.
(b) Quantification of phosphatase activity for PhoQ1*,PhoQ5*,PhoQ12*, and PhoQ14*, as in Fig. 2c. Lines indicate mean ± s.d. from n = 3 biological replicates.
(c) Phosphotransfer profiles of wild-type PhoQ (top) and three PhoQ* variants, as in Fig. 2d, but for 60 min and 5 min. In each case, the kinase was autophosphorylated and then incubated for 5 or 60 min. individually with each of 27 response regulators from E. coli, and its selected PhoP* variant, followed by SDS-PAGE and autoradiography. The position of the autophosphorylated kinase and the approximate positions of any phosphorylated regulators are indicated by arrowheads on the left. These experiments were repeated independently two times with similar results.
Extended Data Fig. 5. Insulation of E. coli histidine kinases from PhoP* variants.
Phosphotransfer profiles of five histidine kinases endogenous to E. coli: EnvZ (a), RstB (b), CreC (c), CpxA (d), and PhoR (e). In each case, the kinase was autophosphorylated, then incubated for 5 or 60 min. with its cognate response regulator, with wild-type PhoP, or with one of eleven PhoP* variants, and analyzed as in Extended Data Fig. 4. n = 1 independent experiment.
Extended Data Fig. 6. Insulation of PhoQ* variants from E. coli response regulators, with respect to phosphatase activity.
(a) Phosphatase activity of PhoQ was assessed by measuring the decay of phosphorylated response regulators. Twelve E. coli response regulators were selected for their ability to be stably phosphorylated in vitro by a cocktail of six E. coli histidine kinases (CreC, RstA, PhoR, PhoP, EnvZ and CpxA, each at 250 nM). After 2 hours of pre-incubation with radiolabeled ATP and this kinase cocktail, each regulator was combined with 2 mM PhoQ and phosphorylation state of the regulators was measured after 0, 60, and 120 minutes. n = 1 independent experiment.
(b) Phosphatase profiles conducted as in (a) for PhoP* variants. Quantification of wild-type PhoP and PhoP* variant phosphorylation (normalized to t = 0 to display decay) is plotted on the right. n = 1 independent experiment.
(c) Phosphatase profiles conducted as in (a) for PhoQ* variants. n = 1 independent experiment.
(d) Ratio of response regulator phosphorylation between phosphatase profiles with PhoQ* variants (c) and wild-type PhoQ (a,b).
Extended Data Fig. 7. RNA-seq analysis of strains harboring PhoQ*-PhoP* variants.
(a) RNA-seq analysis of strains containing wild-type PhoQ-PhoP or the indicated variant pair, measured after 30 min. with 10 μM or 50 mM Mg2+. Each strain displays a similar Mg2+-limitation induction of three genes (mgtA, mgtL, and mgrB) in the PhoP regulon, as well as the PhoP-dependent reporter gene yfp.
(b) The expression change of each response regulator and histidine kinase gene in E. coli with colors representing the log2 fold change in response to low extracellular Mg2+. Note that rstAB are part of the PhoP regulon and so show changes in transcription following activation of PhoQ and several PhoQ* variants. Otherwise, most two-component pathways show little induction by the wild-type PhoQ-PhoP and the PhoQ*-PhoP* pairs.
(c) The same data as in (b) but with fold change of each variant pair normalized to the fold change seen with wild-type PhoQ-PhoP, where the latter is the geometric mean of two wild-type replicates.
(d) p values of the Z-score calculated for each value in (c). For each gene and each variant, Z-scores represent the deviation of that gene’s variant/wild-type ratio when compared to the distribution of every gene’s variant/wild-type ratio. Using all E. coli genes with reads across multiple samples (n = 3477), p values were calculated with a one-tailed Z-test to identify genes induced more strongly with the variant pairs than with the wild-type pair. The statistical significance threshold after correcting for multiple hypothesis testing is indicated on the color legend encoding p values. None of the other two-component signaling genes in E. coli are significantly induced by the variant PhoQ*-PhoP* pairs tested.
Extended Data Fig. 8. Additional characterization of orthogonal sets of PhoQ*-PhoP* variant pairs.
(a) Reproducibility of replicates for the combinatorial library in Fig. 3a. Correlations between bin frequencies of individual variant pairs measured in independent replicates. R2 values indicate the Pearson correlation coefficients, calculated using all data points (n = 210,319 variant pairs).
(b) Counts of cells sorted into each bin, for each replicate and growth condition.
(c) Functional PhoQ*-PhoP* variants that are orthogonal to wild-type PhoQ and PhoP. The fold-induction values, taken from the matrix in Fig. 3a, measured by Sort-seq for the variant pairs indicated in each row, either together (far right column of the heat map) or with a wild-type protein (middle two columns), compared to the wild-type pair (far left column).
(d) Number of unique sets of various sizes of orthogonal PhoQ*-PhoP* pairs, for various thresholds of activity and cross-talk.
(e) Frequency of each PhoQ* (top) or PhoP* (bottom) variant within the orthogonal sets of various sizes (fold induction > 15, crosstalk < 6).
(f) Phosphatase- and kinase-locked variants of PhoQ were identified by fusing the catalytic DHp-CA domains to the leucine zipper GCN4 at different fusion sites (see Methods).
(g) Phosphatase-locked PhoQ15* is sufficient to suppress nonspecific phosphotransfer from wild-type PhoQ to PhoP15*. (h) Heat map as in Fig. 3a, but restricted to variants that retain an interaction (fold induction > 10) with wild-type PhoQ and PhoP, which are shown in red.
(i-j) Orthogonal sets of PhoQ* and PhoP* variants that, like the set in Fig. 3h, comprise exclusively proteins that retain interactions with the parent PhoQ and PhoP.
(k) Number of unique sets of various sizes of orthogonal PhoQ*-PhoP* pairs in which all variants retain an interaction (fold induction > 10) with wild-type PhoP and PhoQ.
Extended Data Fig. 9. Specificity residues of novel PhoQ*-PhoP* pathways are distinct from all extant two-component signaling interfaces.
(a) Hamming distance was calculated between the 11 specificity residues of each PhoQ*-PhoP* variant pair and the 11 specificity residues of wild-type PhoQ-PhoP, the two-component signaling paralogs in E. coli, or all extant two-component signaling proteins. When comparing two sets, all distances between all members of both sets are plotted.
(b) Force-directed graph representing the sequence space of histidine kinases. Each node represents a single histidine kinase, with the relative positions reflecting the similarity of their interface residues (see Methods). Colored nodes highlight specific sets of kinases as indicated in the legend.
Extended Data Fig. 10. Global insulation of the AQ4*-P4* chimeric signaling pathway.
(a) Phosphotransfer profile of PhoQ4*. PhoQ4* was autophosphorylated and then incubated for 5 and 60 min. with each of 27 response regulators from E. coli and with PhoP4*, as in Extended Data Fig. 4c. This experiment was repeated independently two times with similar results.
(b-c) RNA-seq analysis, as in Extended Data Fig. 7b-c, of strains expressing AQ4* and either wild-type PhoP or PhoP4* (measured after 30 min. induction with 0 mM or 1 mM trans-zeatin).
(b) PhoP regulated genes yfp, mgtL, and mgrB are induced by trans-zeatin only when AQ4* is paired with PhoP4*.
(c) The expression change of all response regulators and histidine kinases (fold change in response to 10 mM Mg2+ or 1 mM trans-zeatin).
Supplementary Material
Acknowledgements
We thank I. Nocedal, P. Culviner, D. Ding, and A. Podgornaia for helpful discussions. M.T.L. is an Investigator of the Howard Hughes Medical Institute. This work was also supported by a grant from the Office of Naval Research (N000141310074) to M.T.L and C.A.V and by the NIH Pre-Doctoral Training Grant T32GM007287.
Footnotes
Data Availability
Datasets generated during this study have been deposited in GEO. Raw reads and processed Sort-seq analysis of each mutant can be found under the accession numbers GSE120780 (degenerate PhoQ-PhoP library) and GSE120786 (combinatorial library of 79 PhoQ*-PhoP* variants). Raw reads and RPKM for all Escherichia coli genes from RNA-seq are deposited with the accession number GSE128611.
Code Availability
Python scripts for analysis available at https://github.com/mcclune/nature2019.
References
- 1.Alm E, Huang K & Arkin A The evolution of two-component systems in bacteria reveals different strategies for niche adaptation. PLoS Comput Biol 2, e143 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Capra EJ & Laub MT Evolution of two-component signal transduction systems. Annu Rev Microbiol 66, 325–347 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Capra EJ, Perchuk BS, Skerker JM & Laub MT Adaptive mutations that prevent crosstalk enable the expansion of paralogous signaling protein families. Cell 150, 222–232 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zarrinpar A, Park SH & Lim WA Optimization of specificity in a cellular protein interaction network by negative selection. Nature 426, 676–680 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Stiffler MA et al. PDZ domain binding selectivity is optimized across the mouse proteome. Science 317, 364–369 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brentjens RJ et al. CD19-targeted T cells rapidly induce molecular remissions in adults with chemotherapy-refractory acute lymphoblastic leukemia. Sci Transl Med 5, 177ra138 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Riglar DT & Silver PA Engineering bacteria for diagnostic and therapeutic applications. Nat Rev Microbiol 16, 214–225 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Morsut L et al. Engineering Customized Cell Sensing and Response Behaviors Using Synthetic Notch Receptors. Cell 164, 780–791 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bashor CJ, Helman NC, Yan S & Lim WA Using engineered scaffold interactions to reshape MAP kinase pathway signaling dynamics. Science 319, 1539–1543 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Sockolosky JT et al. Selective targeting of engineered T cells using orthogonal IL-2 cytokine-receptor complexes. Science 359, 1037–1042 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skerker JM, Prasol MS, Perchuk BS, Biondi EG & Laub MT Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol 3, e334 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Creixell P et al. Unmasking determinants of specificity in the human kinome. Cell 163, 187–201 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thompson KE, Bashor CJ, Lim WA & Keating AE SYNZIP protein interaction toolbox: in vitro and in vivo specifications of heterospecific coiled-coil interaction domains. ACS Synth Biol 1, 118–129 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boyken SE et al. De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity. Science 352, 680–687 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reinke AW, Grant RA & Keating AE A synthetic coiled-coil interactome provides heterospecific modules for molecular engineering. J Am Chem Soc 132, 6025–6031 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stock AM, Robinson VL & Goudreau PN Two-component signal transduction. Annu Rev Biochem 69, 183–215 (2000). [DOI] [PubMed] [Google Scholar]
- 17.Groban ES, Clarke EJ, Salis HM, Miller SM & Voigt CA Kinetic buffering of cross talk between bacterial two-component sensors. J Mol Biol 390, 380–393 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siryaporn A & Goulian M Cross-talk suppression between the CpxA-CpxR and EnvZ-OmpR two-component systems in E. coli. Mol Microbiol 70, 494–506 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Capra EJ et al. Systematic dissection and trajectory-scanning mutagenesis of the molecular interface that ensures specificity of two-component signaling pathways. PLoS Genet 6, e1001220 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skerker JM et al. Rewiring the specificity of two-component signal transduction systems. Cell 133, 1043–1054 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Podgornaia AI & Laub MT Protein evolution. Pervasive degeneracy and epistasis in a protein-protein interface. Science 347, 673–677 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Casino P, Rubio V & Marina A Structural insight into partner specificity and phosphoryl transfer in two-component signal transduction. Cell 139, 325–336 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Yamada H et al. The Arabidopsis AHK4 histidine kinase is a cytokinin-binding receptor that transduces cytokinin signals across the membrane. Plant Cell Physiol 42, 1017–1023 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Nielsen AA et al. Genetic circuit design automation. Science 352, aac7341 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Mutalik VK et al. Precise and reliable gene expression via standard transcription and translation initiation elements. Nat Methods 10, 354–360 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Ashenberg O, Keating AE & Laub MT Helix bundle loops determine whether histidine kinases autophosphorylate in cis or in trans. Journal of Molecular Biology 425, 1198–1209 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gibson DG et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6, 343–345 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Fowler DM & Fields S Deep mutational scanning: a new style of protein science. Nat Methods 11, 801–807 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Starr TN, Picton LK & Thornton JW Alternative evolutionary histories in the sequence space of an ancient protein. Nature 549, 409–413 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diss G & Lehner B The genetic landscape of a physical interaction. Elife 7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Balakrishnan S, Kamisetty H, Carbonell JG, Lee SI & Langmead CJ Learning generative models for protein fold families. Proteins 79, 1061–1078 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Culviner PH & Laub MT Global Analysis of the E. coli Toxin MazF Reveals Widespread Cleavage of mRNA and the Inhibition of rRNA Maturation and Ribosome Biogenesis. Mol Cell 70, 868–880 e810 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langmead B & Salzberg SL Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eddy SR Accelerated Profile HMM Searches. PLoS Comput Biol 7, e1002195 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jacomy M, Venturini T, Heymann S & Bastian M ForceAtlas2, a Continuous Graph Layout Algorithm for Handy Network Visualization Designed for the Gephi Software. Plos One 9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohlendorf R, Schumacher CH, Richter F & Moglich A Library-Aided Probing of Linker Determinants in Hybrid Photoreceptors. ACS Synth Biol 5, 1117–1126 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Wang B, Zhao A, Novick RP & Muir TW Activation and inhibition of the receptor histidine kinase AgrC occurs through opposite helical transduction motions. Mol Cell 53, 929–940 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marina A, Mott C, Auyzenberg A, Hendrickson WA & Waldburger CD Structural and mutational analysis of the PhoQ histidine kinase catalytic domain. Insight into the reaction mechanism. J Biol Chem 276, 41182–41190 (2001). [DOI] [PubMed] [Google Scholar]
- 39.Miyashiro T & Goulian M Stimulus-dependent differential regulation in the Escherichia coli PhoQ PhoP system. Proc Natl Acad Sci U S A 104, 16305–16310 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.