Abstract
Mitochondrial homeostasis vitally depends on mitophagy, the programmed degradation of mitochondria. The roster of proteins known to participate in mitophagy remains small. We devised here a multidimensional CRISPR/Cas9 genetic screen, using multiple mitophagy reporter systems and pro-mitophagy triggers, and uncover numerous new components of Parkin-dependent mitophagy1. Unexpectedly, we identify the adenine nucleotide translocator (ANT) complex as required for mitophagy in multiple cell types. While pharmacological inhibition of ANT-mediated ADP/ATP exchange promotes mitophagy, genetic ablation of ANT paradoxically suppresses mitophagy. Importantly, ANT promotes mitophagy independently of its nucleotide translocase catalytic activity. Instead, the ANT complex is required for inhibition of the presequence translocase TIM23, leading to PINK1 stabilization, in response to bioenergetic collapse. ANT modulates TIM23 indirectly via interaction with TIM44, known to regulate peptide import through TIM232. Mice lacking ANT1 reveal blunted mitophagy and consequent profound accumulation of aberrant mitochondria. Disease-causing human mutations in ANT1 abrogate binding to TIM44 and TIM23 and inhibit mitophagy. Together, these data identify a novel and essential function for ANT as a fundamental mediator of mitophagy in health and disease.
We carried out a multidimensional CRISPR/Cas9 genome-wide screen to identify new components of the mitophagy machinery. In the predominant pathway for mitophagy, PTEN-induced kinase 1 (PINK1) protein accumulates on damaged mitochondria and recruits the E3 ubiquitin ligase Parkin to target mitochondria for autophagy1,3,4. C2C12 mouse myoblasts were made to stably over-express Parkin, and exposed to two orthogonal mitochondrial stressors (membrane potential uncoupling with CCCP; or suppression of oxidative phosphorylation with a cocktail of inhibitors [OAR: Oligomycin inhibiting complex V; Antimycin A inhibiting complex III; Rotenone inhibiting complex I]), leading to robust mitophagy (Extended Data Fig. 1a,b). Cells were transduced with lentivirus containing Cas9 plus guide RNAs (gRNAs) targeting >20,000 genes 5, treated with either CCCP or OAR, and subjected to one of four flow cytometry-based mitophagy assays: 1. Loss of mitotracker labeling of mitochondrial membrane 6; 2. Loss of ectopically expressed outer membrane-targeted GFP (GFP-Omp25); 3. Loss of ectopically expressed matrix GFP protein (Cox8-GFP); and 4. Altered fluorescence of matrix-targeted mKeima from 440 to 586 as it encounters the low pH environment of the lysosome (Fig. 1a,Extended Data Fig. 1c) 7. High and low fluorescent fractions were sorted and subjected to sequencing. gRNA clones over-represented in the low fraction identified potential mitophagy accelerators, and those in the high fraction potential decelerators. In all seven assays, Pink1 gRNAs scored near the top as mitophagy decelerators, validating the approach (Fig. 1b). An aggregate Z-score metric placed Pink1 gRNAs as the lead decelerator (Fig. 1c). Most proteins reported in the Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Database to regulate mitophagy were identified in the screen (Extended Data Fig. 1d), supporting the robustness of the screen. The complete results of the screen are provided in Table S1.
Fig. 1. Multi-dimensional mitophagy screen reveals that ANT is required for mitophagy.
a, Outline of CRISPR/Cas9 genome-wide genetic screen, using 4 reporter assays and 2 modes of mitophagy induction. b, Most significant hits in each of the 7 screens. Representative previously known genes in open symbols, previously unknown in color; line, median; box, 75–25 percentiles; whiskers, 99–1 percentiles; duplicate experiments. c, Ranked aggregate Z-scores of all genes. Representative previously known in gray, previously unknown in black. d-e, Validation as mitophagy decelerators of the indicated genes, using both a gRNA chosen from the screening library, and an independent non-library gRNA, followed by flow cytometry for mt-mKeima (d, n = 3 biological replicates per gRNA, P values calculated by two-sided unpaired t test relative to NTC) or by Western blotting of mitochondrial proteins in the outer membrane (OMM-Tom20), inner membrane (IMM-ATPB), or matrix (PDH) (e). Similar results were obtained in two biological replicates. For gel source data, see Supplementary Fig. 1. f, Suppression of mitophagy in primary rat neurons. Left: visualization of neuronal mitochondria with TMRE dye. Right: representative image showing coating of mitochondria (labeled with Mito-Snap) with the mitophagy receptor OPTN, indicating active mitophagy. Far right: quantification of cells undergoing active mitophagy; n = 6 (untreated control), 6 (treated control), 4 (ANT1), and 5 biological replicates (ANT2), P values calculated by one-way ANOVA, post-hoc Dunnett’s multiple comparison test, *P < 0.05, **P < 0.01. Scale bar, 5 μm and 0.8 μm. Data are mean ± s.d.
Genes whose perturbation accelerates mitophagy have been less investigated than those that suppress it 1,8,9. Unbiased gene set enrichment analysis (GSEA) of gRNAs that accelerate mitophagy revealed mitochondrial bioenergetics as the most important target class (Extended Data Fig. 2a). 16% of genes encoding mitochondrial proteins were identified, a 3-fold over-representation over all genes (Extended Data Fig. 2b), in particular genes involved in oxidative phosphorylation (OXPHOS) (Extended Data Fig. 2c), consistent with bioenergetic dysfunction being a key promoter of mitophagy. OXPHOS genes were over-represented in each screen (Extended Data Fig. 2d) and markedly so in aggregate (Extended Data Fig. 2e).
GSEA of decelerators of mitophagy revealed a wide range of classes (Extended Data Fig. 2f). Nearly every component of major complexes known to be required for autophagy were identified, including the endosomal sorting complexes required for transport (ESCRT), the homotypic fusion and vacuole protein sorting (HOPS) complex 10-13 (Extended Data Fig. 2g,h), and autophagosome formation (Extended Data Fig. 2i). Variability between screens was higher between assays than between triggers to mitophagy (Extended Data Fig. 2j), despite the dramatically different bioenergetic effects of CCCP and OAR (Extended Data Fig. 2k), suggesting that different forms of bioenergetic collapse converge on a common mitophagy pathway. Autophagy genes scored only in inner membrane and matrix-based screens (Extended Data Fig. 2d), consistent with degradation of the outer membrane requiring different machinery 14. The LC3 receptor Tax1bp1 and its regulatory kinase Tbk1 scored highly (Extended Data Fig. 1d) and were validated with independent gRNAs and orthogonal assays (Extended Data Fig. 3a,b). Deletion of all five LC3 receptors reduced mitophagy levels as much as did Pink1 deletion (Extended Data Fig. 3c), indicating significant redundancy. Co-deletion of Tax1bp1 and Tbk1 had similar synergistic effects (Extended Data Fig. 3d).
The adenine nucleotide transporter (ANT), located on the inner membrane, mediates trans-membrane exchange of ADP and ATP, providing ADP to the mitochondrial ATPase (Complex V). ANT is not known to be involved in mitophagy. Strikingly, both Ant1 (Slc25a4) and Ant2 (Slc25a5) scored highly in our screen (Fig. 1b,c), and were validated with independent gRNAs (Fig. 1d) and by orthogonal assay using Western blotting to detect degradation of mitochondrial components (Fig. 1e,Extended Data Fig. 4a). Deletion of Ant1 or Ant2 in mouse N2A neuroblastoma cells that express endogenous parkin, and deletion of ANT2 in human SH-SY5Y neuroblastoma cells (which only express ANT2), also potently suppressed mitophagy (Extended Data Fig. 4b-c). Finally, siRNA-mediated suppression of ANT1 or ANT2 in primary rat hippocampal neurons profoundly blocked the engulfment of mitochondrial fragments by the autophagy receptor OPTN, a quantifiable marker of mitophagic events in these cells and nonneuronal cell lines15 (Fig. 1f). Thus, we find that ANT is required for parkin-mediated mitophagy in numerous cell types.
These observations were surprising. Blocking ADP/ATP exchange worsens CCCP-induced membrane depolarization (Extended Data Fig. 5a), and would be predicted to accelerate mitophagy rather than decelerate it16,17. Indeed, pharmacological inhibition of ADP/ATP exchange aggravated mitophagy (Fig. 2a,Extended Data Fig. 5b). In contrast, genetic deletion of Ant1 or Ant2 prevented mitophagy, despite worsening the loss of membrane potential after CCCP treatment (Fig. 2b). These data indicated that deletion of Ant1 or Ant2 blocks mitophagy via mechanisms other than blocking ADP/ATP exchange. Consistent with this conclusion, ectopic expression of ANT1 bearing mutations (K43E/R244E) that interfere with binding to ATP 18 and ADP/ATP exchange (Extended Data Fig. 5c,d) rescued mitophagy in cells lacking ANT1 as efficiently as did wildtype ANT1 (Fig. 2c). In contrast, two mutations, A123D and A90D, identified in patients with cardiomyopathy and external ophthalmoplegia, respectively 19, failed to rescue mitophagy (Fig. 2c), despite evidence that ADP/ATP exchange is preserved 16(Extended Data Fig. 5d). Thus, ANT promotes mitophagy independently of its ADP/ATP exchange activity.
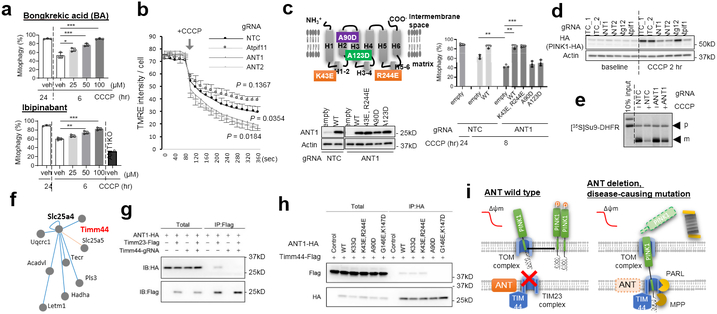
Fig. 2. ANT mediates suppression of TIM23 via TIM44.
a, Inhibition of ADP/ATP transport with the indicated inhibitors accelerates mitophagy, in sharp contrast to genetic deletion of ANT1 (bottom right); n = 3 per group. b, Mitochondrial Δψm is elevated in cells lacking ANT, and reduced in response to CCCP, compared to control cells, consistent with reverse ADP/ATP exchange in low Δψm; n = 3 per gRNA. P values calculated by two-way repeated measures ANOVA relative to NTC. c, Rescue of mitophagy with wild type (WT) ANT and ADP/ATP-binding mutant (K43E/R244E), but not with disease-causing mutants (A90D, A123D). top left: schematic of ANT mutations. Bottom: equivalent expression of constructs. Right: quantification of mitophagy; n = 3 per group. d, PINK1 stabilization by CCCP treatment is abrogated in the absence of ANT1 or 2. e, Suppression of TIM23-mediated import of of [35 S]Su9-DHFR into isolated mitochondria in response to CCCP is impaired in the absence of ANT1 (p=precursor; m=mature). f, Interaction between ANT1 and Timm44 identified in the mitochondrial interactome database XlinkDB. g, Timm44 deletion impairs ANT1 binding with Timm23. h, Mutation of the predicted Timm44 interaction site in ANT1, or disease-causing ANT1 mutation (A90D), abrogate bind to Timm44, while ADP/ATP exchange mutants (K43E/R244E and K33Q) do not. i Model of ANT-mediated suppression of TIM23 in response to mitochondrial bioenergetic compromise. Data are mean ± s.d. P values by one-way ANOVA, post-hoc Tukey test (a,c), *P < 0.05, **P < 0.01, ***P < 0.001. Similar results were obtained in two biological replicates. For gel source data, see Supplementary Fig. 1 (c-e,g,h).
Loss of ANT prevented mitophagy induced by oxidative stress (H2O2) and OXPHOS poisons, both PINK1-dependent processes, but not by hypoxia or nutrient deprivation, both PINK1-independent processes (Extended Data Fig. 5e), suggesting that ANT acts upstream of PINK. In the absence of mitochondrial damage, PINK1 is constitutively degraded by the cytosolic proteasome, after being partly imported into mitochondria through the TIM23 complex, leading to the proteolytic removal in the matrix of its transmembrane domain, and allowing its subsequent release to the cytoplasm20. When mitochondrial damage occurs, TIM23-mediated translocation is inhibited, causing PINK1 to retain its transmembrane domain, incorporate into the outer membrane, escape degradation, auto-phosphorylate, and recruit Parkin to the mitochondria1,20,21. In the absence of ANT1 or ANT2, however, PINK1 stabilization in response to CCCP was prevented (Figs. 2d,Extended Data Fig. 5f). PINK stabilization could be rescued by wild-type or K43E/R244E ANT1, but not by A90D and A123D mutants (Extended Data Fig. 5g). PINK1 autophosphorylation (Extended Data Fig. 5h), mRNA expression (Extended Data Fig. 5i), or protein translation (Extended Data Fig. 5j) were not affected by the absence of ANT. PARL or OMA1, proteases known to be involved in PINK1 processing22,23 (Extended Data Fig. 5k), and general autophagy (Extended Data Fig. 5l), were similarly unaffected. In sharp contrast, the CCCP-triggered suppression of TIM23-mediated protein translocation into the matrix (Extended Data Fig. 5m), and the cleavage in the matrix of imported Su9-DHFR precursor (p) to the mature (m) form (Fig. 2e) were markedly blunted in cells lacking Ant1 or Ant2, indicating that ANT is required for suppression of TIM23. The loss of ANT did not affect the integrity of TIM23 and TOM mega-complexes (Extended Data Fig. 6a). ANT1 and 2 interact with TIM23 (Extended Data Fig. 6b-c), as does the ATP-binding mutant K43E/R244E, but not the disease-causing mutants A90D and A123D (Extended Data Fig. 6d). K43E/R244E ANT1 rescued the suppression of TIM23 in cells lacking ANT1, but the A90D and A123D mutants did not (Extended Data Fig. 6e). Together, these data demonstrate that ANT mediates suppression of TIM23 translocase in response to mitochondrial bioenergetics collapse, and it does so by interacting with TIM23, independently of its activity as an ADP/ATP exchange carrier.
To understand how ANT interacts with TIM23, we probed a mitochondrial interactome database24, and identified TIM44 as a leading candidate interactor with ANT1 (Fig. 2f). TIM44 is a matrix-side component of the TIM23 complex that controls translocation of polypeptides across TIM23 2. TIM44 co-immunoprecipitated with ANT1 and 2 (Extended Data Fig. 6b,f). Knockout of TIM44 abrogated TIM23 binding to ANT1 (Fig. 2g), indicating that TIM44 likely mediates the TIM23/ANT1 interaction. Like loss of ANT1, loss of TIM44 blunted both mitophagy (Extended Data Fig. 6g) and PINK1 accumulation (Extended Data Fig. 6h). A mutant of ANT1 unable to bind to TIM44 (ANT1 G146E/K147D; Fig. 2h) failed to rescue mitophagy in cells lacking ANT1 (Extended Data Fig. 6i). Importantly, the disease-causing mutant A90D also disrupted interaction with TIM44 (Fig. 2h). Conversely, a mutant of TIM44 unable to bind to ANT1 (TIM44 K282D; Extended Data Fig. 6j) failed to rescue mitophagy in cells lacking TIM44 (Extended Data Fig. 6k), despite preserved ability to bind to TIM23 itself (Extended Data Fig. 6l). Together, these data demonstrate that ANT1 interacts with TIM44, a known regulator of TIM23-mediated protein translocation2, and that the interaction of ANT1 with TIM44 is required for the ability of ANT1 to interact with TIM23, to suppress TIM23-mediated protein translocation in response to bioenergetics collapse, and to promote mitophagy (Fig. 2i).
To test if ANT is required for mitophagy in vivo, we examined mice lacking Ant125. Mitochondria isolated from heart or brain of Ant1KO mice accumulated remarkably less Pink1 and Parkin than mitochondria from littermate controls, despite equivalent or higher amounts of Pink1 and Parkin transcription (Fig. 3a,Extended Data Fig. 7). Similarly, expression in skeletal muscle of mitochondria-targeted tandem GFP-mCherry, a method to quantify lysosome-targeted mitochondria as red-only punctae26,27, revealed a profound defect in mitophagy in Ant1KO mice (Fig. 3b). Muscle mitophagy was rescued by re-expression of wild-type ANT1 or ANT1 bearing a K33Q mutation found in patients with mild muscular disease despite complete loss of ADP/ATP exchange capacity (Extended Data Fig. 5d, and ref 28), but was not rescued by expressing the A90D mutation, found in patients with severe disease despite normal ADP/ATP exchange (Fig 3c, and Extended Data Fig. 5d). Consistent with defective mitophagy, heart and skeletal muscle of Ant1KO mice revealed marked accumulation of mitochondria, as shown by mitochondrial DNA (Fig. 3d) and protein (Fig. 3e), electron microscopy (Fig. 3f), and deep coloring of skeletal muscle29 (Fig. 3g). Importantly, the expression of nuclear-encoded mitochondrial genes is unaffected in Ant1KO animals (Fig. 3d), indicating that the accumulation of mitochondria occurs without mitochondrial biogenesis. Moreover, the accumulated mitochondria are profoundly abnormal, with swelling, loss of cristae architecture and density (Fig. 3f), and accumulated DNA mutations30-32, all consistent with a loss of quality control and clearance caused by a loss of mitophagy. These numerous abnormalities lead to marked cardiac hypertrophy 29, and loss of normal contractile function (Fig. 3h).
Fig. 3. ANT is required for mitophagy in vivo.
a, Blunted mitophagy in heart and brain of Ant1KO animals, illustrated by reduced coating of mitochondria by p62, PINK1, and Parkin. b, Blunted mitophagy in skeletal muscle of Ant1KO animals, shown by intramuscular transfection of mitoQC-plasmid; Line, mean; n = 4 mice, 4 fields (WT) and 4 mice, 50 fields (ANT1 knockout). Scale bar, 10 μm. c, Rescue of mitophagy with wild type (WT) ANT1 and mutant lacking ADP/ATP exchange activity (K33Q), but not with disease-causing mutant (A90D); Line, mean; n = 3 mice, 24 fields (WT), 3 mice, 16 fields (Luc), 3 mice, 23 fields (K33Q) and 3 mice, 21 fields (A90D). Scale bar, 10 μm. d, Accumulation of mitochondrial DNA (right) in heart (top) and muscle (bottom) of Ant1KO animals despite absence of nuclear-encoded biogenesis (left); n = 4 per group. e-g, Accumulation of mitochondrial proteins (e) and of disorganized and aberrant mitochondria (f) in heart, and deep red coloring of mitochondria in skeletal muscle (g) of Ant1 KO animals. Scale bar, 2 μm. Similar results were obtained from three mice in each group. h, Dilated cardiomyopathy in Ant1KO animals: heart weights (top left), and sample images (top right) and quantification (bottom) of echocardiography; n = 7 per group. LVID: left ventricle internal diameter systolic (s) and diastolic (d); LVPW: left ventricle posterior wall; EF: ejection fraction. i, Echocardiography of patient bearing homozygous loss-of-function ANT1 mutations. Left: systole. Right: diastole. j, Electron micrographs of endomyocardial biopsy from same patient as in (i). Localized distension of the outer membrane (blue arrows) with apparent release of mitochondrial matrix content (green) into the cytoplasm. Scale bar, 1 μm. Data are mean ± s.d. (a,e). P values by two-sided unpaired t test (a,d,h), by one-way ANOVA, post-hoc Tukey test (b), *P < 0.05, **P < 0.01, ***P < 0.001. For gel source data, see Supplementary Fig. 1.
Finally, to evaluate the consequences of loss of ANT1 in human cardiac tissue, we identified a patient bearing homozygous loss-of-function mutations in ANT1 (frameshift at position 522, p.Y174fs) in the Penn Medicine BioBank (PMBB). The patient developed severe non-ischemic cardiomyopathy with biventricular failure, with left ventricular ejection fraction of ~15% (normal: ~55%) (Fig. 3i). Endomyocardial biopsy revealed profound mitochondrial abnormalities, including swelling, loss of cristae, and diffuse burst mitochondria with compromised outer membranes and release of mitochondrial components into the cytoplasm, all consistent with loss of mitophagy and mitochondrial quality control (Fig. 3j).
In summary, we show that ANT is critically required for mitophagy in cell culture and in vivo; that ANT interacts with TIM23 and is required for suppression of TIM23-mediated protein translocation and subsequent PINK1 stabilization; that ANT modulates TIM23 via interaction with TIM44, a known regulator of TIM23 polypeptide import; and that strikingly this role for ANT in mitophagy is independent of its ADP/ATP exchange capacity. ANT is uniquely positioned, as a proton gradient-dependent carrier of ADP/ATP, to sense bioenergetic failure from either proton gradient collapse (e.g. with CCCP) or failure to carry oxidative phosphorylation (e.g. with OAR). Mutations in ANT1 cause diseases ranging from cardiomyopathy (A123D) to progressive external ophthalmoplegia (A90D), but the underlying mechanisms are still debated. These diseases typically manifest a high burden of mitochondrial DNA mutations 31,33, suggesting a failure of adequate mitochondrial quality control. In addition, there is poor correlation between ADP/ATP exchange capacity and disease severity34. Our data thus suggest the notion that defects in mitophagy, rather than ADP/ATP exchange, may drive the pathophysiology of ANT1 diseases.
METHODS
Cells.
C2C12 myoblast, SH-SY5Y neuroblastoma cells, and N2A neuroblastoma cells were cultured at 37°C with 5% CO2 in Dulbecco’s modified Eagle medium (DMEM, invitrogen) containing 10% fetal bovine serum (HyClone) and penicillin/streptomycin (100U/ml, Invitrogen).
Mice.
All mouse experiments were performed according to procedures approved by the University of Pennsylvania Institutional Animal Care and Use Committees. Male Ant1−/− and Ant1+/+ mice (C57BL/6JEiJ) were fed a standard diet ad libitum. Mice were maintained on a 13:11 h light–dark cycle. Mitophagy was assessed with mito-QC plasmid injection in male 12-16 week-old mice, and echocardiography was performed in male 8-9 month-old mice.
Antibodies.
The following primary antibodies were used: mouse anti-HA (clone 6E2, CST, 1:2000), rabbit anti-HA (clone C29F4, CST, 1:1000), anti-HA (HRP Conjugate, clone C29F4, CST, 14031S, 1:5000), anti-DYKDDDDK Tag (clone D6W5B, 14793S, CST, 1:5000), anti-Flag (clone M2, sigma, F3165, 1:5000), anti-Tom20 (FL-145, Santa Cruz Biotechnology, 1:2000 for immunoblotting or 1:200 for immunostaining), anti-Tom70 (Proteintech, 1:1000), anti-ATPB (clone 3D5, Abcam, 1:1000), anti-ATPA (7H10BD4F9, Invitrogen, 1:2000), anti-Pyruvate dehydrogenase E2/E3bp (clone 13G2AE2BH5, Abcam, 1:1000), anti-beta actin (clone 13E5, CST, 1:2000), anti-ANT1 (Sigma, SAB4300887, 1:1000), anti-ANT1 (Sigma, SAB2108761, 1:1000), anti-ANT2 (Sigma, HPA046835, 1:1000), anti-Timm23 (Abcam, ab116329, 1:1000), anti-Timm23 (Proteintech, #11123, rabbit, 1:1000), anti-Timm22 (Proteintech, 14927-1-AP, rabbit, 1:1000), anti-VDAC1 (CST, #4866S, rabbit, 1:2000), anti-Pink1 (Caymanchem, #10006283, rabbit, 1:500), anti-Parkin (Santa Cruz, PRK8, sc-32282 HRP, mouse, 1:100), anti-p62 (CST, #5114S, rabbit, 1:2000), anti-OPA1 (BD, #612607, mouse, 1:5000), anti-PGAM5 (Abcam, ab126534, rabbit, 1:1000), anti-Tomm40 (Proteintech, #18409, rabbit, 1:5000), anti-GAPDH (CST, #5174S, rabbit, 1:5000), anti-Tubulin (CST, #2128S, rabbit, 1:5000), anti-beta actin (CST, #4970S, rabbit, 1:2000), anti-pan actin (CST, #8456S, rabbit, 1:5000). We also used the following secondary antibodies: anti-mouse-IgG or anti-rabbit-IgG antibody conjugated to horseradish peroxidase (CST, 1:5000) for immunoblotting, and anti-rabbit-IgG antibody conjugated to Alexa Fluor 594 (Life Technologies, 1:2000) for immunostaining.
Plasmids.
Individual gRNAs were cloned into lentiCRISPR v2 (Addgene 52961) or lentiGuide-Puro (Addgene 52963)35 and cDNAs were cloned in pLenti (Addgene 22255), pMSCV (Clontech), or pcDNA4TO (Addgene 60914). Drug resistance genes in lentiGuide, pLenti, and pMSCV were replaced by Neomycin, or Zeocin resistant genes. The resulting plasmid was verified by sequencing. pMD2.G (Addgene 12259) and psPAX2 (Addgene 12260) were used for lentiviral packaging. pMD2.G and gag/pol (Addgene 14887) were used for retroviral packaging.
Gene-specific cDNA rescue.
A gRNA-resistant version of ANT1 was amplified by the primer encoding silent mutations within the gRNA recognition sites. This cDNA was cloned into pMSCV-neo for cell experiments and pAAV-CMV for in vivo mitophagy measurements.
Virus production.
To produce lentivirus for the screen, we used the GeCKOv2mouse library in the lentiGuide-Puro vector35. 10cm plates of 70% confluent Lenti-X 293T (Clontech) cells were transfected with 9 μg of the plasmid library, 6 μg of PAX2 vector and 3 μg pMD2.G using Fugene HD (Promega) according to the manufacturer’s instructions. Supernatant was collected after 48 hrs and then spun for 10 min at 4°C at 3000 rpm and then filtered with a 0.45μm 0.45uM low protein-binding filter (SFCA) and concentrated by ultracentrifugation for 2 hrs at 24,000 rpm using SW28Ti. The virus was aliquoted and frozen at −80°C. The titer of the virus was determined by using C2C12 cells followed by puromycin selection. For other viruses, 6 well plate of 70% confluent Lenti-X 293T cells were transfected with 1.5 μg of transfer vector, 0.5 μg pMD2.G and 1.0 μg of PAX2 for lentivirus or 1.0 μg of gag/pol for retrovirus using Fugene HD. Supernatant was collected after 48 hrs and frozen at −80°C.
CRISPR library cell preparation.
C2C12 cells were first infected with lentiviruses prepared from pLenti hParkin and pFUGW spCas9 (Addgene 52962) with 8 μg/mL polybrene. 24 hrs after infection, cells were treated with 2mg/ml Hygromycin (Mediatech) and 10 μg/mL Blastcidin (Invitrogen) for 4days and maintained 1 weak without drugs. Several single cell clones were developed and tested the ability to induce mitophagy. The well-induced clone was expanded and additionally infected with lentivirus prepared from pLenti Cox8-GFP, pLenti GFP-Omp25, or pLenti Cox4-mKeima, and then GFP or mKeima positive cells were sorted with narrow signal range. These cells were subsequently infected with the genome-wide GeCKO lentiviral library (Addgene 1000000053) at a MOI of ~0.5 (to yield ~100 cells per gRNA) with 8μg/mL polybrene in 6-well plates, with 0.6×106 cells infected per well. Infected cells were treated with 2.5 μg/mL puromycin (Clontech) for 4 days, and the cells were expanded for an additional 6 days without puromycin. All virus infection was performed by centrifugation at 1000 g for 90 min, then the medium was removed, and cells were incubated in fresh medium for 24 hrs.
Mitophagy induction.
To induce mitophagy, mitochondrial depolarization or inhibition of OXPHOS was induced in human Parkin-expressing cells. In the screen, cells were treated with 13 μM CCCP (Sigma) or cocktail of 1.0 μg/mL Oligomycin (Sigma), 1.0 μg/mL Antimycin A (Sigma) and 100 nM Rotenone (Sigma) for 16 hrs for mild induction of mitophagy to harvest enough cells in low induction population. In other experiments, cells were treated with 15 or 20 μM CCCP (MCE or Sigma) or cocktail of 3 μg/mL Oligomycin and 3 μg/mL Antimycin A.
FACS.
Cell sorting was performed by BD FACSJazz and BD Influx (BD Biosciences). After mitophagy induction, cells were harvested by trypsinization, washed and resuspened with FACS buffer (PBS containing 2% FBS and 20mM HEPES). For sorting by MitoTracker, cells were incubated with 100 nM MitoTracker Green FM (CST) for 20min after mitophagy induction. Cells were sorted into two bins (low and high). The bin boundaries were guided by our observations with or without mitophagy induction in MitoTracker-guided sorting. In other sorting, there was no clear separation between high and low population. Two bins were set to sort top 25-30% and bottom 25-30%. Measurements of lysosomal mt-mKeima were made using dual-excitation ratiometric pH measurements at 488 (pH 7)- and 532 (pH 4)-nm lasers with 670/30-nm and 58¾0-nm emission filters, respectively. In each library, ~40 million cells were sorted and ~10 million cells were collected in each bin. For non-pooled CRISPR knockout experiments with individual gRNAs, BD LSR II (BD Biosciences) and Attune NxT Flow Cytometer (Thermo Fisher Scientific) were used. Analysis was done with FlowJo (Treestar).
DNA purification, library preparation, and sequencing.
Genomic DNA from sorted cells was purified and eluted in 500 μl H2O using Blood and Cell Culture DNA Midi kit (Qiagen) according to the manufacturer’s instruction. More than 100 μg of DNA were typically obtained from the analyses. We performed 3-step PCR reactions using TaKaRa Ex Taq and PrimeSTAR HS (Clontech). First PCR was the enrichment of gRNA sequence from 100 μg extracted gDNA in 1 ml reaction. Second and 3rd PCR were to attach Illumina sequencing adapters and sample barcodes with 1-5 μl from the prior PCR. PCR cycling conditions were 2min at 98°C; followed by 25 cycles (1st PCR) or 7 cycle (2nd and 3rd PCR) of 10s at 98°C, 15s at 55°C, 60s at 72°C; and a final 2 min extension at 72°C. The final PCR product was run on a 2% agarose gel and the right size fragment was extracted with QIAquick Gel Extraction kit (Qiagen). Samples were sequenced on a HiSeq 2500 (Illumina). gRNA reads were mapped to a reference index of all GeCKO mouse library gRNA sequences using the short read aligned STAR 36 and the following parameters (--outFilterMultimapNmax 41 --outFilterMismatchNmax 4 --outFilterScoreMinOverLread 0 --outFilterMatchNminOverLread 0 --outFilterMatchNmin 17). Uniquely mapped reads were counted for each gRNA for each sample.
Screen statistical analysis.
To determine the enrichment of gRNAs between treatment and control, we used the Z score method describes in Parnas et.al 37. Briefly count data was quantile normalized per replicate to adjust for differences in library sizes. Replicates were summarized on the gRNA guide level using a geometric mean and a fold change was computed between the treatment and the control. To summarise at the gene level the mean of the top four ranked gRNAs was taken for positive regulators, and the bottom four ranked gRNAs for negative regulators. Empirical P values were calculated by randomly assigning gRNAs to genes and false discovery rates (FDR) were assessed using the Benjamini and Hochberg method.
Individual gRNAs validation.
The best gRNA from the GeCKO library and 1 or more non-library gRNAs were cloned into lentiGuide-Puro plasmid (Addgene 52963). Non-library gRNA was designed at http://crispr.mit.edu/. Two non-targeting control in GeCKO library were used as a negative control.
Mitochondrial isolation and immunoprecipitation.
Cells were harvested in PBS and washed using isolation buffer (250 mM Sucrose, 10 mM HEPES: pH 7.4, 1 mM EGTA). Cells in isolation buffer were homogenized on ice using a Glass Pestle Tissue Grinder (Corning). Cell debris was removed by centrifugation at 1000 g for 10 min. Mitochondria were collected by centrifugation at 10000 g for 10 min.38 Freshly isolated mitochondria were washed in isolation buffer and protein concentrations were determined by bicinchoninic acid protein assay. Standard immunoprecipitation experiments were performed with 0.5–1 mg mitochondria and analysed by SDS-PAGE and immunoblot analysis. Briefly, isolated mitochondria from control cells (empty vector) and cells expressing HA tagged proteins were solubilised in digitonin-containing buffer (1% [w/v] digitonin, 20 mM Tris, pH 7.5, 0.1 mM EDTA, 50 mM NaCl, 10% [w/v] glycerol) supplemented with 1x complete protease (Roche) at 2 mg/mL end over end on a rotary wheel for 30–60 min at 4°C. The mitochondrial lysate was cleared by centrifugation at 16,000 g at 4°C for 10 min and diluted in solubilisation buffer (20 mM Tris, pH 7.5, 0.1 mM EDTA, 50 mM NaCl, 10% [w/v] glycerol, 1x complete protease inhibitor) so that a final digitonin concentration of 0.5% was achieved. The cleared lysates were applied to pre-equilibrated anti-HA Magnetic Beads (Thermo Fisher Science) and incubated at 4°C for 60 min under mild agitation. The anti-HA beads and bound proteins were washed x 3 in digitonin-containing buffer (0.1% digitonin [w/v], 20 mM Tris, pH 7.5, 0.1 mM EDTA, 50 mM NaCl, 10% [w/v] glycerol, 1x complete protease inhibitor). Bound proteins were eluted with 2x Laemmli Sample Buffer, then electrophoretically separated using SDS-PAGE.39
Phos-tag SDS-PAGE and immunoblotting.
To detect phosphorylated proteins, SDS-PAGE with Phos-tag acrylamide gels (Wako chemicals) were used. After electrophoresis, Phos-tag acrylamide gels were washed using transfer buffer with 5 mM EDTA for 10 min with gentle shaking and then replaced with transfer buffer without EDTA for 10 min according to the manufacturer’s protocol. Proteins were transferred to PVDF membranes and detected by the indicated antibodies using standard immunoblotting procedures.40
ΔΨ measurements.
Mitochondrial membrane potential was determined by TMRE according to previous report.41 Briefly, TMRE measurements were performed by LSM 710 confocal laser scanning microscopy (Carl Zeiss Inc.) with the application of live time-series program and using a 20× objective with low-resolution (512 × 512) and attenuated laser power (1%) to avoid photobleaching. Cells were incubated with 40 nM TMRE in Tyrode’s buffer (TB: 145 mM NaCl, 5 mM KCl, 10 mM glucose, 1.5 mM CaCl2, 1 mM MgCl2, and 10 mM HEPES; adjust pH to 7.4 with NaOH) for 30 min prior to imaging and treatment. Use region of interest (ROI) tool from the LSM program to select the areas. Select ROIs from the entire cell body in imaged cells to measure the TMRE intensity per cell. Calculate the average fluorescence intensities from whole cell bodies for all imaged cells for each time point.
In vivo mitophagy measurements.
Mice (ANT1 knockout and wild type) were anesthetized with 1-4% isoflurane. Small incision was made on the skin to expose TA muscle. 25ug of mitoQC plasmid dissolved in sterile saline was intramuscularly injected with 33G needle attached to Hamilton syringe. For the co-transfection experiment 20ug of mitoQC and 5ug of pcDNA3-Luciferase or ANT1 (WT, K33Q or A90D) were injected. Skin incision was then sutured and electric pulse (voltage, 50 V/cm; pulse duration, 100 ms; frequency of pulses, 1 Hz) was applied by two pin electrodes. A train of total eight pulses (four pulses, then an additional four pulses after switching the position of the electrodes) was delivered. Mice were rested for more than 10days. Then mice were sacrificed and TA muscles were fixed with 4% PFA for overnight. For cryoprotection, fixed muscles were then submerged in 30% sucrose/PBS until the tissue pieces sunk. Tissues were then mounted with O.C.T. compound and frozen in 2-methyl butane chilled with liquid N2. Tissue blocks were longitudinally sectioned and mounted with dapi containing mounting media. Transfected fibers were searched by GFP signal and confocal images were taken. Each fiber was analyzed by Image J. Briefly confocal image was spitted into green and red channels and bright objects in each channel were identified by local thresholding algorithm (Phansalker) and binary image of respective objects was created. Red objects touching with green objects were subtracted. Size filtering (<0.15 um^2) was applied to filter out the noise. Remaining red objects were counted by particle analysis. Number of red objects was normalized by fiber area. Over 40 fibers from 5 different mice (8-10 fibers per mouse) were analyzed in each genotype.
Electron Microscopy.
For EM studies, gastrocnemius was isolated, and red and white fibers were immediately dissected and fixed with 2.5% (vol/vol) glutaraldehyde, 2.0% (vol/vol) paraformaldehyde in 0.1 M sodium cacodylate buffer, pH 7.4, overnight at 4 °C. EM studies were performed by JEM-1010 microscope at University of Pennsylvania Electron Microscopy Resource Laboratory.
Constructs and siRNA.
The following constructs were used: Mito-DsRed2 (provided by T. Schwarz, Harvard Medical School, Boston) was recloned into a SNAP-Tag vector, and pEGFP-OPTN (provided by I. Dikic, Goethe University, Frankfurt). The following Dharmacon siRNA oligos were used: Rat ANT1/Slc25a4 siRNA ON-TARGETPlus SMARTpool, Rat ANT2/Slc25a5 siRNA ON-TARGETPlus SMARTpool, and fluorescent ON-TARGETPlus Nontargeting siRNA with 5’ Cy5.
Primary Hippocampal Culture.
Rat hippocampal neurons, E18, were obtained from the Neurons R Us Culture Service Center through the Penn Medicine Translational Neuroscience Center at the University of Pennsylvania. Cells were plated at density of 250,000 neurons per 35 mm glass-bottom MatTek dish that were precoated with 0.5 ml/ml poly-L-lysine for 24 hrs. Neurons were maintained in maintenance media (MM; neurobasal supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin, 2 mM GlutaMax, and 2% B27) at 37 C in a 5% CO2 incubator. AraC (5 μM) was added the day after plating.
Neuronal Mitophagy Assay.
At 7 DIV, siRNA (45 pmol) and DNA constructs (0.3 μg Mito-SNAP and 0.4 pEGFP-OPTN) were transfected using Lipofectamine 2000 and incubated for 36-48 hrs. Prior to imaging, neurons were treated for 2 hrs with control or 3 nM Antimycin A and labeled with 2 μM SNAP Blue 430 (Promega). 1.5 hrs into the incubation, cells were quickly washed twice with MM to remove excess SNAP label and incubated for 30 min in control and Antimycin A media plus 2.5 nM TMRE. For live-cell imaging, neurons were placed in HibernateE supplemented with 2% B27 and 33 mM D-glucose; 3 nM Antimycin A was added to the imaging media for treated conditions. Neurons were imaged in an environmental chamber at 37 C on a Perkin Elmer UltraView Vox spinning disk confocal on a Nikon Eclipse Ti Microscope with a 100× 1.49 NA oil-immersion objective and a Hamamatsu EMCCD C9100-50 camera driven by Volocity. Z-stacks of the soma were acquired at 150 nm step-size. For mitophagy analysis, the number of clearly defined OPTN rings around damaged, spherical mitochondria were counted for each cell. The percentage of cells with mitophagy events were quantified and graphed in GraphPad Prism. Statistical analysis was carried on data sets that were performed from more than four independent preparations. One-way ANOVA tests with Dunnett’s post hoc test was performed were values were compared to the Control siRNA with 2 hr Antimycin A treatment.
Penn Medicine BioBank.
The Penn Medicine BioBank (PMBB) is a longitudinal genomics and precision medicine study, in which participants consent to linkage of biologic specimens with electronic health record data. Currently, more than 50,000 participants are enrolled. The study was approved by the Institutional Review Board of the University of Pennsylvania.
ADP/ATP exchange rate.
ADP/ATP exchange rate by magnesium green (Invitrogen) was determined in permeabilized myotubes as described previously 42. Briefly, C2C12 myoblasts were transfected with WT, K33Q, K43E, A90D, V289M, A114P ANT1 adenovirus. C2C12 myoblasts were induced differentiation into myotubes by replacing culture medium with differentiation medium containing DMEM and 2% house serum. Myotubes were processed on the 6th day for subsequent experiments. Cells were washed once and resuspended in a buffer containing 8 mM KCl, 110 mM K-gluconate, 10 mM NaCl,10 mM Hepes, 10 mM KH2PO4, 0.005 mM EGTA, 10 mM mannitol, 0.5 mM MgCl2. Digitonin and MgG 5 K+ salt were subsequently added. MgG fluorescence was recorded at a 0.33-Hz acquisition rate by using 505- and 535-nm excitation and emission wavelengths, respectively. Mitochondrial phosphorylation was started by the addition of 2 mM ADP. At the end of each experiment, minimum fluorescence was measured after the addition of 5 mM EDTA, and maximum fluorescence was measured by the addition of 10 mM MgCl2. ADP-ATP exchange rates were calculated by linear regression of the magnesium green fluorescence calibrated and converted to ATP appearing in the medium42.
In vitro transcription and translation.
Prior to import into purified mitochondria, [35S]-methionine labeled precursor protein were generated with TNT Quick Coupled Transcription/Translation kit (Promega) and TRAN35S-LABEL, Metabolic Labeling Reagent (MP Biomedicals).
Import of radiolabeled proteins into mitochondria.
Ten micrograms of C2C12 cell mitochondria were incubated in import buffer (20 mM HEPES pH 7.4, 220 mM mannitol, 70 mM sucrose, 1 mM ATP, 0.5 mM magnesium acetate, 20 mM sodium succinate, 5 mM NADH) for 5 mins at 25°C. The membrane potential was disrupted by treatment with 20 μM CCCP (MCE). After adding 10ul precursor, culture for 10 mins at 25°C. Samples were transferred to ice and add 500ul cold homogenization buffer. Trypsin was added to a final concentration of 10 μg/ml, and samples were incubated on ice for an additional 15 mins. The trypsin digestion was terminated by adding soybean trypsin inhibitor to a final concentration of 250 μg/ml. Mitochondria were re-isolated by centrifugation at 10,000g for 5 mins. Samples were resuspended in SDS sample buffer and resolved by SDS–PAGE.
Blue native PAGE (BN-PAGE).
Mitochondria were isolated from indicated C2C12 myoblasts and were lysed in BN lysis buffer (20 mM HEPES pH 7.4, 50 mM NaCl, 2.5 mM MgCl2, 10% glycerol, 0.1 mM EDTA, 0.5 mM PMSF, 1.5% or 0.5% digitonin; AG scientific) at a concentration of 5 mg/ml for 30 min on ice. After a centrifugation at 21,000g for 10 min the mitochondrial extract was separated on a NativePAGE 3-12% Bis-Tris Gels (Invitrogen) or 4-15% Precast-Glgel 4-15% Native PAGE Gels (Sangon Biotech). Proteins were transferred to a PVDF membrane and detected by immunoblot using the indicated antibodies.
Statistical analyses.
All data were expressed as mean ± s.d. No statistical methods were used to predetermine sample size. Sample size was based on experimental feasibility and sample availability. Randomization was not applicable to mouse studies due to genotype-dependent analyses. Samples were processed in random order and mitophagy signal count, heart weight and echocardiography were measured in a blinded manner. Comparisons between the two groups were analysed using the two-sided unpaired t test. One-way ANOVA followed by Tukey’s post hoc test, or post-hoc Dunnett’s multiple comparison test was used for multiple group comparisons. Two-way repeated measures ANOVA was used for the comparisons in TMRE membrane potential experiments; P<0.05 was considered statistically significant.
Reporting summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Data availability
All data generated or analyzed during this study are included in this published article (and its supplementary information files).
Extended Data
Extended Data Figure 1. CRISPR library screening for PINK1-Parkin mediated mitophagy.
a,b, Mitophagy was induced in parkin and mt-mKeima expressing cells by treatment with a mitochondrial membrane potential uncoupler CCCP or a cocktail of suppressors of oxidative phosphorylation (OAR: Oligomycin, Antimycin A, and Rotenone). Mitophagy was analyzed by flow cytometry for mt-mKeima (a) and western blotting for mitochondrial protein in the outer membrane (Tom20), inner membrane (ATPB), or matrix (PDH) (b). c, Representative gate setting of cell sorting for each of the four indicated mitophagy assays: 1. Loss of MitoTracker labeling of mitochondrial membrane; 2. Loss of ectopically expressed outer membrane-targeted GFP (GFP-Omp25); 3. Loss of ectopically expressed matrix GFP protein (Cox8-GFP); and 4. Altered fluorescence of matrix-targeted mKeima. d, Genes in the KEGG mitophagy pathway. Genes identified as mitophagy accelerators or decelerators in CRISPR knockout, defined as Z-score > 1.5 in at least one screen, are indicated in green and red, respectively. Diameter of each circle is proportional to the Z-score of the indicated gene. Similar results were obtained in two biological replicates (a-c). For gel source data, see Supplementary Fig. 1.
Extended Data Figure 2. Integration of seven mitophagy screens.
a, Gene set enrichment analysis of mitophagy accelerators. The top 1% of genes in aggregate Z-score were analyzed using ToppGene Suite31,43. Representative functional categories and Bonferroni-corrected P values are shown. b, Proportion of genes encoding mitochondrial proteins annotated in MitoCarta2.044 in the top 1 % of mitophagy accelerators and decelerators (left), and percentage of MitoCarta2.0 member genes identified as being either accelerators (green) or decelerators (red) of mitophagy (right). c, Five functional classes of proteins, based on MitoCarta2.0 annotations, were present in the top 1% of mitophagy accelerators. The representation of each class within the top 1% was compared to its representation in MitoCarta2.0 via a two-tailed Fisher’s exact test (FET). d, Box-and-whisker plots of most significant mitophagy accelerator hits in each of the 7 screens; line, median; box, 75-25 percentiles; whiskers (blue dots), 99–1 percentiles. Genes involved in oxidative phosphorylation (OXPHOS) are indicated in yellow. Pathway enrichment was calculated using a Kolmogorov-Smirnov (K-S) test.e, GSEA enrichment plot for OXPHOS (top) and ranked aggregate Z-scores of all genes. OXPHOS genes are indicated in yellow (bottom). f, Gene set enrichment analysis of mitophagy decelerators analyzed as (a). g-i, Genes in the KEGG endosomal sorting complexes required for transport (ESCRT) (g), homotypic fusion and vacuole protein sorting (HOPS) (h), and autophagosome (i) pathways. Genes identified as mitophagy accelerators or decelerators are indicated in green and red, respectively. Diameter of each circle is proportional to the Z-score of the indicated gene. j, Principle component analysis biplot summarizing variation across the seven screens based on cumulative z-scores of the top 100 genes, displayed as arrows. Autophagy-related genes are indicated. k, Mitochondrial membrane potential assessed by flowcytometry for TMRE is disrupted in CCCP treatment, but is increased in a cocktail of suppressors of oxidative phosphorylation (OAR: Oligomycin, Antimycin A, and Rotenone). Similar results were obtained in two biological replicates.
Extended Data Figure 3. Essential LC3 receptors for mitophagy in C2C12 mouse myoblasts.
a,b, Validation as mitophagy decelerators of the indicated LC3 receptor gene gRNAs, using one library gRNAs and one non-library gRNAs and using the mt-mKeima assay (a) or Western blotting of mitochondrial proteins in the outer membrane (Tom20 and Tom70), inner membrane (ATPB), or matrix (PDH) (b); n = 3 biological replicates per gRNA, P values calculated by two-sided unpaired t test relative to NTC_1. Mitophagic degradation of mitochondrial inner membrane and matrix proteins, but not outer membrane proteins, were blocked by gRNA targeting Tax1bp1 or Tbk1, consistent with the notion that ubiquitinated outer membrane proteins can be degraded by the ubiquitin proteasome system. Similar results were obtained in two biological replicates. c,d, LC3 receptor redundancy and TBK1 contribution. The indicated gRNAs were transduced singly or in combination by lentivirus infection, followed by analysis by flow cytometry for mt-mKeima. Multiple gRNAs were superimposed on cells already targeted by Tax1bp1 gRNA (c) or by Tbk1 gRNA (d), as indicated; n = 3 biological replicates per gRNA. P values calculated by one-way ANOVA, post-hoc Tukey test, *P < 0.05, **P < 0.01, ***P < 0.001. Data are mean ± s.d.
Extended Data Figure 4. The adenine nucleotide transporter (ANT) is required in Parkin-mediated mitophagy.
a, Impaired mitophagy in the absence of ANT is confirmed by flux analysis using a lysosome inhibitor, Bafilomycin A (1μM). Similar results were obtained in two biological replicates. b-c, Inhibition of mitophagy by CRISPR-mediated deletion of the indicated genes in murine N2A (b) and human SH-SY5Y (c) neuroblastoma cell lines. Representative flow tracings are shown on left, and quantification on right; n = 3 biological replicates per gRNA, P values calculated by two-sided unpaired t test relative to NTC_1. Data are mean ± s.d.
Extended Data Figure 5. The adenine nucleotide transporter (ANT) is required in PINK1 stabilization.
a, Inhibition of ADP/ATP worsens the loss of membrane potential in response to CCCP. TMRE fluorescence intensity following treatment with CCCP was analyzed by confocal laser scanning microscopy with the application of live time-series program. Cells were pretreated with control and Bongkrekic acid; n = 4 biological replicates per group. P values calculated by two-way repeated measures ANOVA. b, Mitophagy was induced by OXPHOS inhibitors (Antimycin A and Oligomycin) in the presence of indicated concentration of ADP/ATP transport inhibitor, Bongkrekic acid, followed by flow cytometry for mt-mKeima; n = 3 biological replicates per group, P values calculated by one-way ANOVA, post-hoc Tukey test, **P < 0.01. c, Genetic inhibition of ADP/ATP worsens the loss of membrane potential in response to CCCP. ANT1 knockout cells were rescued by human wild-type (WT) or mutant ANT1; n = 3 biological replicates per group, P values calculated by two-way repeated measures ANOVA relative to NTC. d, ADP/ATP exchange rate is impaired in ADP/ATP-binding mutants (K33Q, K43E), but not in disease-causing mutants (A90D, V289M); n = 3 biological replicates per group, P values calculated by two-sided unpaired t test relative to WT. e, Loss of ANT impairs PINK1-dependent mitophagy induced by oxidative stress, but does not impair PINK1-independent mitophagy caused by hypoxia or starvation; n = 3 biological replicates per gRNA, P values calculated by two-sided unpaired t test relative to NTC. f, PINK1 accumulation in mitochondria is impaired in cells lacking ANT. Cells bearing gRNA targets to the indicated genes were transduced with PINK1-GFP, followed by treatment with CCCP versus control, and then immunostained using anti-Tom20 antibody (red). GFP fluorescence is shown in green, and merged signal in yellow. Scale bar, 20 μm. g, PINK1 stabilization by CCCP treatment is preserved in WT ANT and ADP/ATP-binding double mutant (K43E/R244E), but not in known disease-causing mutants (A90D, A123D). h, Phosphorylation of PINK1 after CCCP treatment is preserved in the absence of ANT1 or 2. i-j, PINK1 transcription (i) and translation (j) are not changed by loss of ANT; n = 4 biological replicates per group, P values calculated by one-way ANOVA. k, The activities of PINK1-cleaving protease, PARL and OMA1, are not changed by loss of ANT. l, General autophagy flux is preserved in the absence of ANT; n = 3 biological replicates per group, P values calculated by two-sided unpaired t test relative to NTC. m, Suppression of TIM23-mediated protein translocation in response to CCCP treatment is impaired in the absence of ANT1 or 2, as shown by import of Su9-GFP into intact cells; n = 3 biological replicates per gRNA, P values calculated by two-sided unpaired t test relative to NTC, scale bar, 20 μm. Data are mean ± s.d. Similar results were obtained in two biological replicates (f-h,j,k). For gel source data, see Supplementary Fig. 1.
Extended Data Figure 6. The adenine nucleotide transporter (ANT) mediates closure of TIM23 via TIM44.
a, Deletion of Ant1 or Ant2 does not affect expression of TIM and TOM proteins (right) or destabilize TIM and TOM complexes, as assessed by blue native PAGE (left). b-c, ANT1 and 2 bind to Timm23 and Timm44, as assessed by co-immunoprecipitation (b) and blue native PAGE (c). ANT and TIM23 complex is marked with an asterisk. d, WT ANT1 and the ADP/ATP binding double mutant (K43E/R244E) bind to TIM23 component Timm23, while disease-causing mutants (A90D, A123D) do not. e, Closure of TIM23 in response to CCCP treatment is impaired in the presence of disease-causing mutants (A90D, A123D), but is preserved in the presence of ADP/ATP binding double mutant (K43E/R244E), as shown by import of Su9-GFP to mitochondria; n = 3 biological replicates per group, P values calculated by two-sided unpaired t test relative to empty. Scale bar, 40 μm. f, ANT1 binds to both Timm23 and Timm44. g, Mitophagy is impaired in cells lacking Timm44; n = 3 biological replicates per gRNA, P values calculated by two-sided unpaired t test relative to NTC_1. h, PINK1 stabilization by CCCP treatment is abrogated in the absence of Timm44. i, Rescue of mitophagy with wild type (WT) ANT and ADP/ATP exchange mutants (K33Q, K43E/R244E), but not with known disease-causing mutants (A90D, A123D) and Timm44-binding site mutant (G146E/K147D). top left: schematic of ANT and sites of mutations. Bottom: Western blotting demonstrating equivalent expression of ANT constructs. Right: quantification of mitophagy; n = 3 biological replicates per group, P values calculated by one-way ANOVA, post-hoc Tukey test, **P < 0.01, ***P < 0.001. j, Mutation of the predicted ANT1 interaction site in Timm44 abrogates binding to ANT1. k, Rescue of mitophagy with WT Timm44, but not with binding site mutant (K282D); n = 3 biological replicates per group, P values calculated by one-way ANOVA, post-hoc Tukey test, **P < 0.01, ***P < 0.001. l, Mutation in Timm44 of the ANT1 interaction site does not abrogate Timm44 binding to Timm23. Data are mean ± s.d. Similar results were obtained in two biological replicates (a-d,f,h-j,l). For gel source data, see Supplementary Fig. 1.
Extended Data Figure 7. The adenine nucleotide transporter (ANT) is required for mitophagy in vivo, independently of transcriptional regulation.
Equivalent amounts of Pink1 and higher Parkin transcription in ANT1_KO heart and skeletal muscle (SM); n = 4 per group, P values calculated by two-sided unpaired t test. Data are mean ± s.d.
Supplementary Material
Acknowledgments:
We thank the staff of the Penn Medicine Biobank including JoEllen Weaver, David Birtwell, Heather Williams, Paul Baumann, and Marjorie Risman, as well as the Regeneron Genetics Center (RGC). The Penn Medicine Biobank was funded by a gift from the Smilow family and by the Penn Cardiovascular Institute and the Perelman School of Medicine. AH was supported by the Uehara Memorial Foundation, Mochida Memorial Foundation for Medical and Pharmaceutical Research and JSPS Overseas Research Fellowships. SW was supported by a fellowship from the ADA (1-16-PDF-117) and Toyobo Biotechnology Foundation. CSE was supported by a Hanna H. Gray Fellowship from the Howard Hughes Medical Institute. ELFH was supported by the NIH NINDS (R37 NS060698). DCW was supported by the NIH (NS021328, OD010944, MH108592, MH110185) and DOD (W81XWH-16-1-0401) ZA was supported by the NIH (HL094499, DK107667) and the AHA (Established Investigator Award).
Footnotes
Competing interests: The authors declare no competing interests.
References
- 1.Youle RJ & Narendra DP Mechanisms of mitophagy. Nat Rev Mol Cell Biol 12, 9–14 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ting SY, Yan NL, Schilke BA & Craig EA Dual interaction of scaffold protein Tim44 of mitochondrial import motor with channel-forming translocase subunit Tim23. Elife 6(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matsuda N, et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol 189, 211–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saito T & Sadoshima J Molecular mechanisms of mitochondrial autophagy/mitophagy in the heart. Circ Res 116, 1477–1490 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cottet-Rousselle C, Ronot X, Leverve X & Mayol JF Cytometric assessment of mitochondria using fluorescent probes. Cytometry A 79, 405–425 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Katayama H, Kogure T, Mizushima N, Yoshimori T & Miyawaki A A sensitive and quantitative technique for detecting autophagic events based on lysosomal delivery. Chem Biol 18, 1042–1052 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Hoshino A, et al. Cytosolic p53 inhibits Parkin-mediated mitophagy and promotes mitochondrial dysfunction in the mouse heart. Nat Commun 4, 2308 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Hasson SA, et al. High-content genome-wide RNAi screens identify regulators of parkin upstream of mitophagy. Nature 504, 291–295 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McEwan DG, et al. PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol Cell 57, 39–54 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Lee JA, Beigneux A, Ahmad ST, Young SG & Gao FB ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr Biol 17, 1561–1567 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Hammerling BC, et al. A Rab5 endosomal pathway mediates Parkin-dependent mitochondrial clearance. Nat Commun 8, 14050 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takahashi Y, et al. An autophagy assay reveals the ESCRT-III component CHMP2A as a regulator of phagophore closure. Nat Commun 9, 2855 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshii SR, Kishi C, Ishihara N & Mizushima N Parkin mediates proteasome-dependent protein degradation and rupture of the outer mitochondrial membrane. J Biol Chem 286, 19630–19640 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong YC & Holzbaur EL Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc Natl Acad Sci U S A 111, E4439–4448 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawamata H, Tiranti V, Magrane J, Chinopoulos C & Manfredi G adPEO mutations in ANT1 impair ADP-ATP translocation in muscle mitochondria. Hum Mol Genet 20, 2964–2974 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chevrollier A, Loiseau D, Reynier P & Stepien G Adenine nucleotide translocase 2 is a key mitochondrial protein in cancer metabolism. Biochim Biophys Acta 1807, 562–567 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Di Marino D, Oteri F, della Rocca BM, D'Annessa I & Falconi M Mapping multiple potential ATP binding sites on the matrix side of the bovine ADP/ATP carrier by the combined use of MD simulation and docking. J Mol Model 18, 2377–2386 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Clemencon B, Babot M & Trezeguet V The mitochondrial ADP/ATP carrier (SLC25 family): pathological implications of its dysfunction. Mol Aspects Med 34, 485–493 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Nguyen TN, Padman BS & Lazarou M Deciphering the Molecular Signals of PINK1/Parkin Mitophagy. Trends Cell Biol 26, 733–744 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Okatsu K, et al. PINK1 autophosphorylation upon membrane potential dissipation is essential for Parkin recruitment to damaged mitochondria. Nat Commun 3, 1016 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin SM, et al. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol 191, 933–942 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sekine S, et al. Reciprocal Roles of Tom7 and OMA1 during Mitochondrial Import and Activation of PINK1. Mol Cell 73, 1028–1043 e1025 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Schweppe DK, et al. Mitochondrial protein interactome elucidated by chemical cross-linking mass spectrometry. Proc Natl Acad Sci U S A 114, 1732–1737 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graham BH, et al. A mouse model for mitochondrial myopathy and cardiomyopathy resulting from a deficiency in the heart/muscle isoform of the adenine nucleotide translocator. Nat Genet 16, 226–234 (1997). [DOI] [PubMed] [Google Scholar]
- 26.McWilliams TG, et al. mito-QC illuminates mitophagy and mitochondrial architecture in vivo. J Cell Biol 214, 333–345 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McWilliams TG, et al. Basal Mitophagy Occurs Independently of PINK1 in Mouse Tissues of High Metabolic Demand. Cell Metab 27, 439–449 e435 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.King MS, et al. Expanding the phenotype of de novo SLC25A4-linked mitochondrial disease to include mild myopathy. Neurol Genet 4, e256 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morrow RM, et al. Mitochondrial energy deficiency leads to hyperproliferation of skeletal muscle mitochondria and enhanced insulin sensitivity. Proc Natl Acad Sci U S A 114, 2705–2710 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Esposito LA, Melov S, Panov A, Cottrell BA & Wallace DC Mitochondrial disease in mouse results in increased oxidative stress. Proc Natl Acad Sci U S A 96, 4820–4825 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaukonen J, et al. Role of adenine nucleotide translocator 1 in mtDNA maintenance. Science 289, 782–785 (2000). [DOI] [PubMed] [Google Scholar]
- 32.McManus MJ, et al. Mitochondrial DNA Variation Dictates Expressivity and Progression of Nuclear DNA Mutations Causing Cardiomyopathy. Cell Metab (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fontanesi F, et al. Mutations in AAC2, equivalent to human adPEO-associated ANT1 mutations, lead to defective oxidative phosphorylation in Saccharomyces cerevisiae and affect mitochondrial DNA stability. Hum Mol Genet 13, 923–934 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Thompson K, et al. Recurrent De Novo Dominant Mutations in SLC25A4 Cause Severe Early-Onset Mitochondrial Disease and Loss of Mitochondrial DNA Copy Number. Am J Hum Genet 99, 860–876 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanjana NE, Shalem O & Zhang F Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11, 783–784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parnas O, et al. A Genome-wide CRISPR Screen in Primary Immune Cells to Dissect Regulatory Networks. Cell 162, 675–686 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoshino A, et al. Oxidative post-translational modifications develop LONP1 dysfunction in pressure overload heart failure. Circ Heart Fail 7, 500–509 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Kang Y, et al. Tim29 is a novel subunit of the human TIM22 translocase and is involved in complex assembly and stability. Elife 5(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okatsu K, Kimura M, Oka T, Tanaka K & Matsuda N Unconventional PINK1 localization to the outer membrane of depolarized mitochondria drives Parkin recruitment. J Cell Sci 128, 964–978 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Joshi DC & Bakowska JC Determination of mitochondrial membrane potential and reactive oxygen species in live rat cortical neurons. J Vis Exp (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kawamata H, Starkov AA, Manfredi G & Chinopoulos C A kinetic assay of mitochondrial ADP-ATP exchange rate in permeabilized cells. Anal Biochem 407, 52–57 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen J, Xu H, Aronow BJ & Jegga AG Improved human disease candidate gene prioritization using mouse phenotype. BMC Bioinformatics 8, 392 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Calvo SE, Clauser KR & Mootha VK MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res 44, D1251–1257 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its supplementary information files).