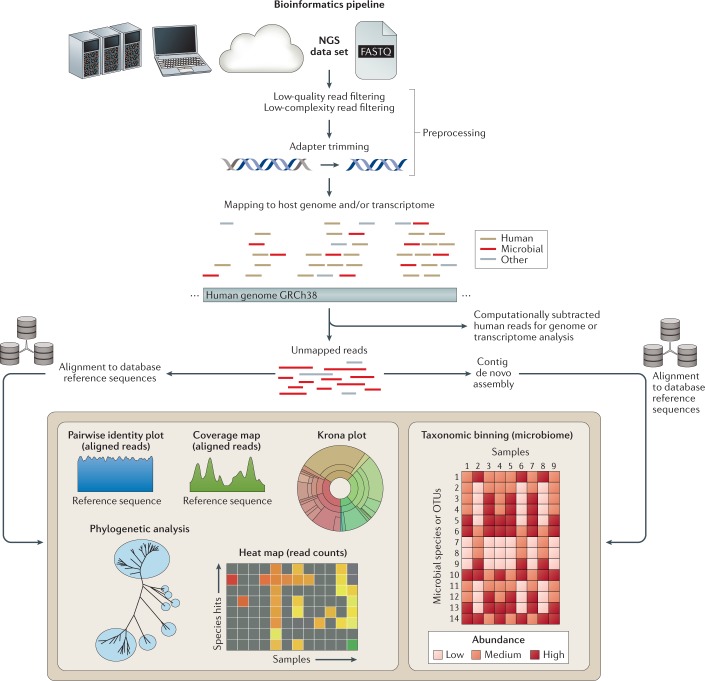
Fig. 4. A typical metagenomic next-generation sequencing bioinformatics pipeline.
A next-generation sequencing (NGS) data set, generally in FASTQ or sequence alignment map (SAM) format, is analysed on a computational server, portable laptop or desktop computer or on the cloud. An initial preprocessing step consists of low-quality filtering, low-complexity filtering and adaptor trimming. Computational host subtraction is performed by mapping reads to the host (for example, human) genome and setting aside host reads for subsequent transcriptome (RNA) or genome (DNA) analysis. The remaining unmapped reads are directly aligned to large reference databases, such as the National Center for Biotechnology Information (NCBI) GenBank database or microbial reference sequence or genome collections, or are first assembled de novo into longer contiguous sequences (contigs) followed by alignment to reference databases. After taxonomic classification, in which individual reads or contigs are assigned into specific taxa (for example, species, genus and family), the data can be analysed and visualized in a number of different formats. These include coverage map and pairwise identity plots to determine how much of the microbial genome has been recovered and its similarity to reference genomes in the database; Krona plots to visualize taxonomic diversity in the metagenomic library; phylogenetic analysis to compare assembled genes, gene regions or genomes to reference sequences; and heat maps to show microorganisms that were detected in the clinical samples. OTU, operational taxonomic unit.