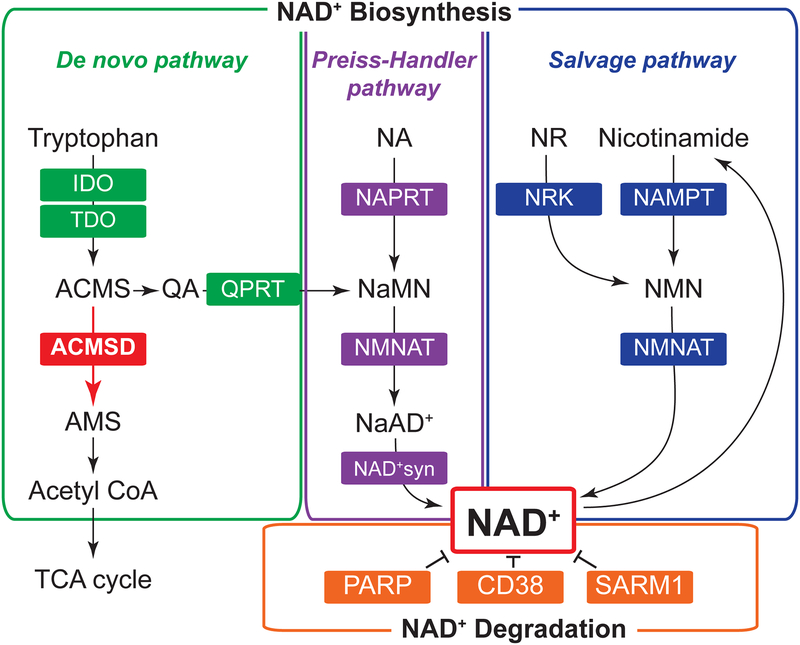
Figure 1. NAD+ biosynthetic and degradative enzymes.
In mammals, nicotinamide adenine dinucleotide (NAD+) homeostasis is regulated by multiple enzymes in de novo biosynthetic pathway starting from tryptophan, Preiss-Handler and salvage biosynthetic pathways starting from niacin (nicotinamide, nicotinic acid [NA], nicotinamide riboside [NR]), and degradative pathway. A new study by Palzer [5] identified alpha-amino-beta-carboxy-muconate-semialdehyde decarboxylase (ACMSD) as an important regulator of NAD+ metabolism. ACMSD coverts aminocarboxymuconic semialdehyde (ACMS) into aminomuconic semialdehyde (AMS) in the de novo NAD+ biosynthetic pathway. Increasing ACMSD expression shifts the balance from de novo NAD+ biosynthesis toward acetyl-CoA production, leading to the development of niacin-dependency.
Aberration: IDO, indoleamine-pyrrole 2,3-dioxygenase; TDO, tryptophan 2,3-dioxygenase; QPRT, quinolinate phosphoribosyltransferase; NAMPT, nicotinamide phosphoribosyltransferase; NMNAT, nicotinamide mononucleotide adenylyltransferase; NRK, nicotinamide riboside kinase; NAPRT, nicotinic acid phosphoribosyltransferase; NAD+syn, NAD+ synthase; QA, quinolinic acid; NMN, nicotinamide mononucleotide; NaMN, nicotinic acid mononucleotide; NaAD+, nicotinic acid adenine dinucleotide; PARP, poly(ADP-ribose) polymerase; SARM1, sterile alpha and TIR motif containing 1; TCA cycle, tricarboxylic acid cycle.