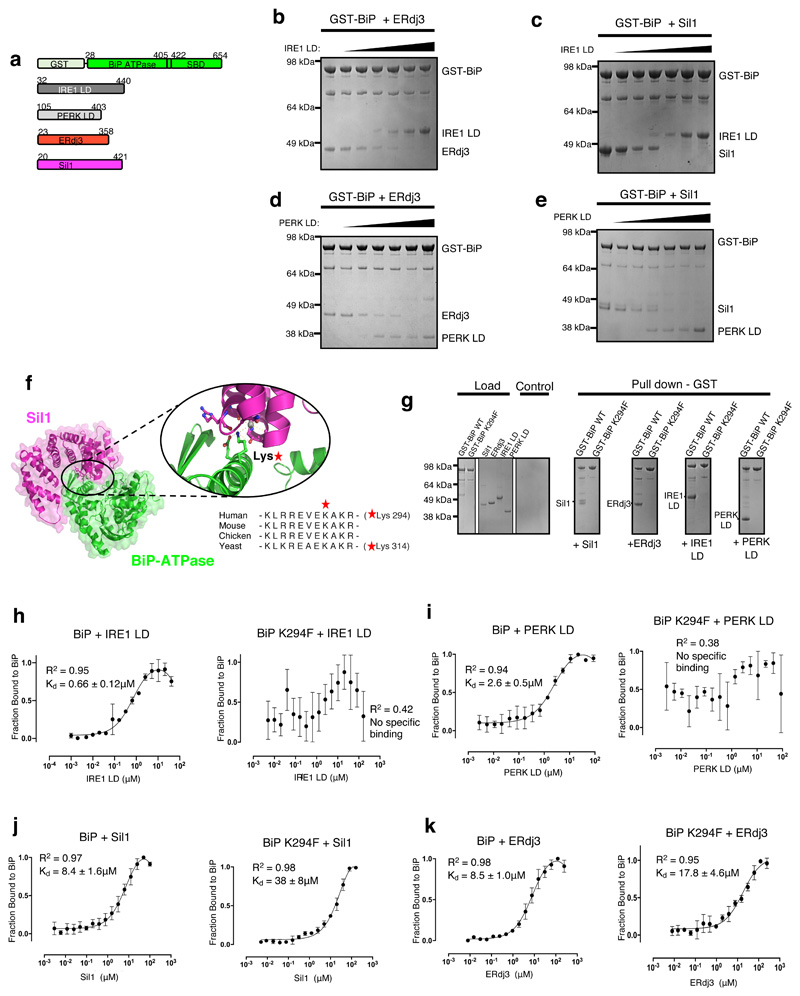
Figure 3. UPR proteins and cochaperones have mutually exclusive binding sites on BiP NBD that are impacted by BiPK294F mutant.
a, Schematic representation of the various constructs used for the pull-down assay, with the numbers denoting the residue number of the protein. b, Competitive pull-down assay in which increasing concentrations of IRE1 LD were added to GST-BiP – ERdj3 chaperone complex. IRE1 LD outcompeted ERdj3 for binding to BiP, suggesting mutually exclusive binding. c, Competitive pull-down assay where GST-BiP - Sil1 complex was immobilized to beads and increasing concentrations of IRE1 LD were added. d, Same as (b), but with PERK LD. e, Same as (c), but with PERK LD. f, Crystal structure of BiP ATPase – Sil1 complex (PDB 3QML), highlighting the interface between interacting molecules. BiP NBD domain residue Lys314 (corresponding to Lys294 in human) mediates key polar contacts between the two molecules. Sequence alignment (right) indicated that this residue is highly conserved across BiP species. g, Pull-down assay to assess interaction of GST- BiPWT or BiPK294F mutant with cochaperones and UPR proteins IRE1 LD and PERK LD. h-k, MST experiments comparing the binding affinity of GST tagged BiPWT and BiPK294F to cochaperones and UPR proteins. Data are mean ± sd from 3 independent experiments. Source Data are available online. BiPK294F mutant disrupted binding to IRE1 LD (h) and PERK LD (i). The BiPK294F mutant displayed greatly reduced binding affinity for Sil1 compared to BiPWT (j). The BiPK294F mutant displayed reduced, but still substantial binding for ERdj3 indicating that ERdj3 may have other contact sites with BiP. Uncropped gel images are available as Source Data. Binding affinities are in Supplementary Table 5.