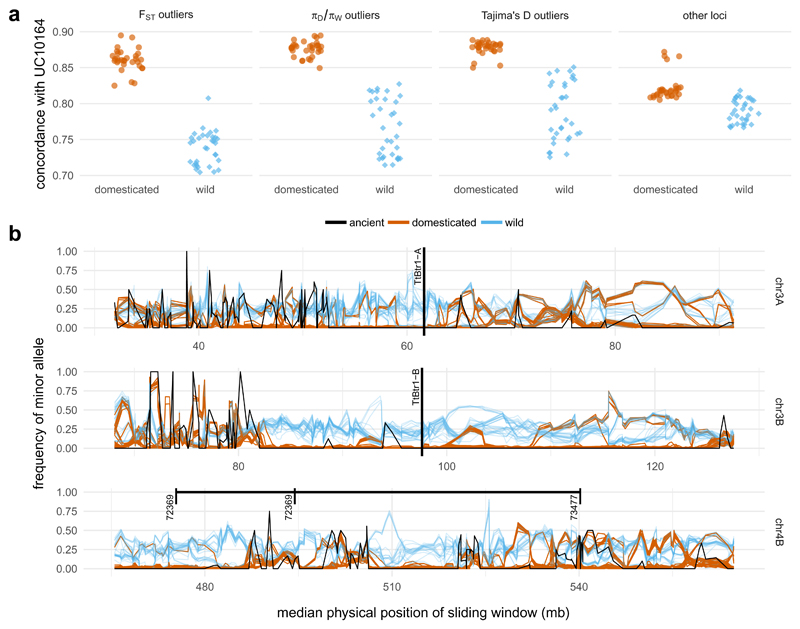
Figure 4. Haplotype sharing between UC10164 and modern emmer wheat within loci associated with selection under domestication.
a, Increased concordance between UC10164 and domesticated accessions (brown) compared to wild accessions (blue), within regions that show putative signatures of selection under domestication, as defined by FST, domesticated/wild nucleotide diversity (πD/πW) and Tajima’s D compared with other non-selected loci. b, The minor allele frequency of all of the samples within 100-SNP sliding windows (moved in 50-SNP intervals) in the regions containing QTLs identified for the following key domestication traits: shattering (chromosomes 3A and 3B) and grain size and seed dormancy (chromosome 4B). The position of putative loss-of-function mutations in TtBr1-A and TtBr1-B is labelled. The range on chromosome 4B shows the maximum extent of plausible positions found for the markers (72369 and 73477) that flank the QTL peak on chromosome 4B. All three cases contain regions in which all of the modern domesticated accessions (brown) and UC10164 (black) have reduced diversity and carry the major allele, which is defined here relative to modern domesticated accessions. Wild accessions are plotted in blue.