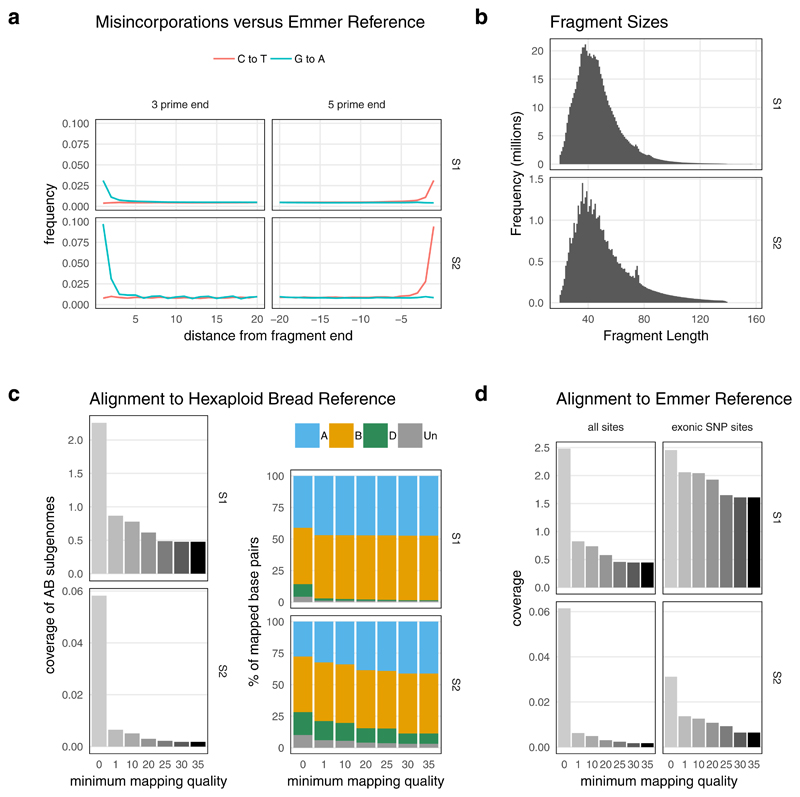
Extended Data Fig. 2. Summary of sequencing data from UC10164 samples S1 and S2.
Summary of sequencing data from UC10164 samples S1 and S2. Panel A shows the C to T and G to A misincorporations of alignments against the emmer wheat reference genome as output by MapDamage. Both samples show a small excess of these misincorporations in the 2bp at each fragment end, as expected for partially UDG treated aDNA libraries. Panel B shows the distribution of fragment sizes after the overlapping paired-end reads were collapsed and adapter sequence was removed using AdapterRemoval. Panel C shows coverage and subgenome representation for different minimum mapping quality scores after these fragments were aligned to the hexaploid bread wheat reference genome. Panel D shows the coverage obtained from alignment to the emmer wheat reference genome, split by minimum mapping quality filters. The exonic SNP sites (at which genotypes are called) have a much lower percentage of ambiguous alignments with low mapping quality scores.