Abstract
Cellular reporters of enzyme activity are based on either fluorescent proteins or small molecules. Such reporters provide information corresponding to wherever inside cells the enzyme is maximally active and preclude minor populations present in sub-cellular compartments. Here we describe a chemical imaging strategy to selectively interrogate minor, sub-cellular pools of enzymatic activity. This new technology confines the detection chemistry to a designated organelle, enabling imaging of enzymatic cleavage exclusively within the organelle. We have thus quantitatively mapped disulfide reduction exclusively in endosomes in C. elegans and identified that exchange is mediated by minor populations of enzymes PDI-3 and TRX-1 resident in endosomes. Impeding intra-endosomal disulfide reduction by knocking down TRX-1 protects nematodes from infection by Corynebacterium diphtheriae, revealing the importance of this minor pool of endosomal TRX-1. TRX-1 also mediates endosomal disulfide reduction in human cells. A range of enzymatic cleavage reactions in organelles are amenable to analysis by this new reporter strategy.
Minor populations of proteins and protein complexes perform critical functions for the cell. For example, a minor population of the epidermal growth factor receptor present on exosomes mediates intercellular communication1. A small fraction of mammalian target of rapamycin, present on lysosomes is responsible for nutrient sensing by the cell2. A minor population of the KDEL receptor present in the Golgi apparatus performs the critical function of retrieving ER-resident proteins from the Golgi apparatus3. Small molecules that function as fluorescent reporters of enzymatic activity use highly specific and rapid detection chemistries. However, the cleaved probe molecules diffuse throughout the entire cell and thus, location-specific information on protein activity is lost4,5. Alternatively, genetically engineered protein tags provide spatial information6,7, however, one can only study the major population of the protein of interest. Thus, it is extremely challenging to selectively address the activity of a minor enzyme population in situ. Here, using DNA nanodevices targeting to sub-cellular organelles we describe a new strategy to exclusively study the activity of a minor population of enzyme in live cells. We demonstrate proof of concept by selectively mapping enzyme-mediated disulfide reduction within endosomes, despite thiol-disulfide exchange occurring rampantly throughout the cell.
Thiol disulfide exchange occurs mainly in the cytosol, mitochondria and the endoplasmic reticulum, to correctly fold disulfide-bridged proteins8,9. Targeted disulfide reduction of specific proteins changes their conformation, thus triggering signalling cascades10. For example, disulfide reduction of C-terminal SRC kinase results in kinase activation leading to cell proliferation and cancer11. However, a small proportion of thiol-disulfide exchange occurs in endocytic organelles. Endosomal disulfide reduction is indispensable to degrading endocytosed proteins and pathogenic material as well as for antigen cross-presentation12,13. In fact, several pathogens exploit this chemistry in the endosome to infect host cells14.
Thiol-disulfide exchange is generally studied using small molecule probes that are either cell-permeable or or do so post endocytosis55,15. Disulfide exchange in the cytosol then enhances probe fluorescence. While highly specific to thiol-disulfide exchange, such probes cannot report on organellar disulfide exchange. Intra-endosomal disulfide reduction was first established in cell lysates post-treatment with radio-labelled, reducible substrates as probes, analysed as a function of time16,17. However, spatial information is inaccessible to these biochemical methods. While responsive to small thiols such as glutathione, protein-based redox probes cannot report on enzyme catalysed disulfide reduction as the disulfide bond in the protein probe is sterically hindered18.
DNA has proven to be a versatile and engineerable biological scaffold for quantitative imaging in living systems19. Functional DNA motifs such as aptamers can recognize a range of analytes, and enabled diverse sensing applications in vitro20. In fact, several detection chemistries have been deployed to ratiometrically image analytes in living systems by leveraging the precise stoichiometry of DNA hybridization to incorporate a reference fluorophore in a well-defined stoichiometry21. Molecular trafficking motifs can be integrated on DNA nanodevices to localize then within organelles, enabling chemical mapping22,23. DNA nanodevices attached to diverse endocytic ligands can target specific endocytic pathways19. We therefore reasoned that by integrating a fluorescent reporter sensitive to thiol-disulfide exchange onto an organelle targetable DNA nanodevice, we would be able to exclusively study endosomal disulfide reduction.
New ratiometric reporters of disulfide exchange
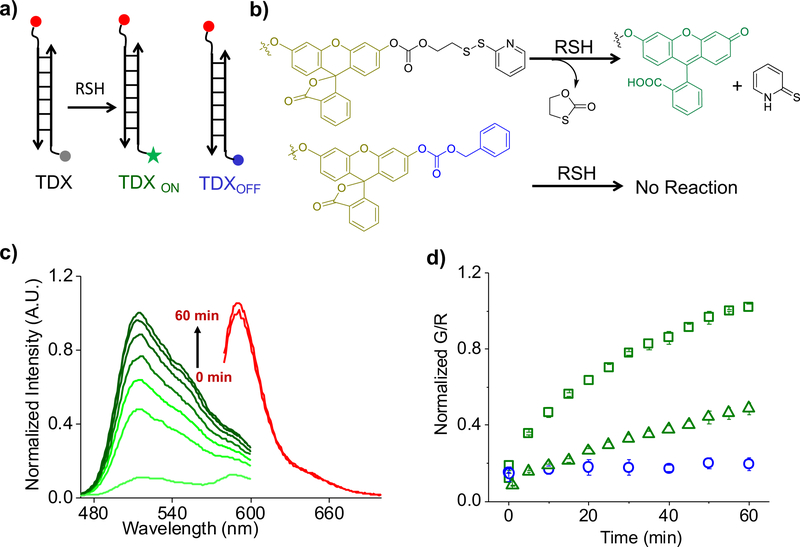
We created a DNA-based, ratiometric reporter consisting of three modules, each with a distinct function (Fig. 1a). The first is a sensing module, consisting of a reaction centre that fluoresces upon thiol-disulfide exchange (Fig. 1a, λem = 520 nm). The second comprises a normalizing module, a rhodamine dye (Fig. 1a, λem= 590 nm) whose fluorescence properties are redox and ion insensitive. The third module comprises a targeting functionality, namely, a DNA duplex comprising two oligonucleotides O-Azide and O-Rhodamine Red X (SI Table 1) that serves two purposes. The first is to display the sensing and normalizing modules in a precise, 1:1 stoichiometry. The second is to target the entire assembly to the scavenger receptor-mediated endocytic pathway24. All three functions are integrated with stoichiometric precision by hybridizing DNA strands bearing each of these functionalities.
Figure 1: Design and in vitro Characterization of TDX reporter.
a) Structure of the TDX reporter (left): the sensing module (grey) is a caged fluorescein dye conjugated with a disulfide moiety, the normalizing module (red) is a thiol-insensitive rhodamine fluorophore, and the targeting module is a DNA duplex (black lines). The reporter (TDX) undergoes thiol disulfide exchange to give highly fluorescent TDXON. TDXOFF (right) is a constitutively off version of TDX where fluorescein is caged by a benzyloxycarbonate group non-responsive to thiols (blue). b) Working principle of sensing module of TDX in presence of reduced thiols. c) Fluorescence emission spectra of sensing module (green, G) and normalizing module (red, R) of TDX in 5 mM glutathione at pH=7.2 as a function of time. d) Ratio of fluorescence intensities (G/R) of TDXOFF (blue open circle, pH=7.2) and TDX in presence of 5 mM glutathione as a function of time at pH=7.2 (green, open square), pH=6.0 (green, open triangle). Error bar indicates the mean of three independent experiments ± s.e.m
The working principle of the sensing module of the disulfide exchange reporter comprises a fluorescein derivative whose fluorescence is caged using carbonate linker (Fig. 1b)4. The other end of the carbonate linker bears a thiopyridyl group via a disulfide bond (Fig.1b, SI scheme 1). Disulfide reduction results in the deprotection of the fluorescein moiety leading to high fluorescence at 520 nm.
To make a quantitative reporter system for disulfide reduction suited to the high autofluorescence encountered in vivo, we made two more nanodevices. One of these, TDXON, comprises the DNA duplex attached to fluorescein (Fig. 1a). TDXON is obtained by completely reducing TDX. As TDX might also turn on due to background hydrolysis of the carbonate linker, we made TDXOFF in which fluorescein is caged by a non-reducible benzyloxycarbonate moiety (Fig. 1a–1b, SI scheme 2). Thus, TDXOFF reports non-specific hydrolysis and reveals the specificity of disulfide exchange reported by TDX. The synthesis and characterization of TDX, TDXON and TDXOFF are described in the supplementary information (SI Scheme 1–2 & SI Fig. 1a).
In the presence of glutathione (GSH, 5 mM) at pH 7.2, TDX fluorescence at 520 nm (G, fluorescein) increases with time, while the fluorescence of the normalizing module, at 590 nm (R, rhodamine), remains constant (Fig. 1c). Fig. 1d shows the G/R ratio of emission intensities as a function of time, which reveals that the reaction is 80% complete in 30 min. Importantly, under comparable conditions, TDXOFF showed no increase in G/R ratio, indicating that the fluorescence increase observed in TDX is due to disulfide exchange (Fig. 1d, SI Fig. 1b).
TDX detects disulfide exchange in late endosome in vivo
Although genetically encoded redox reporters can be applied to study redox in the cytosol18,25 they cannot be used in organelles due to the acidic lumenal pH in organelles. Particularly in Caenorhabditis elegans, the glutathione redox potential varies in different tissues and cell types, yet there is no information available at the sub-cellular level26. We therefore sought to deploy the TDX reporter system to map intra-endosomal thiol disulfide exchange along the endolysosomal pathway in C. elegans.
DNA nanodevices selectively label coelomocytes in C. elegans by targeting scavenger receptor-mediated endocytosis24 Upon injection of 2 μM TDX into the pseudocoelom of wild type (N2) nematodes, fluorescence images of live worms were acquired in G and R channels as a function of time. For each time point we constructed ratiometric maps of G/R intensities represented in pseudocolour (Methods, Fig. 2a). We observed that the G/R ratio was maximal at 20 minutes post injection and remained almost constant thereafter (Fig. 2a–b). A similar experiment using TDXOFF, gave a G/R ratio that remained unchanged (Fig. 2b, SI Fig. 2). This indicates that the increase in G/R ratio shown by TDX is due to thiol-disulfide exchange and not from esterolysis.
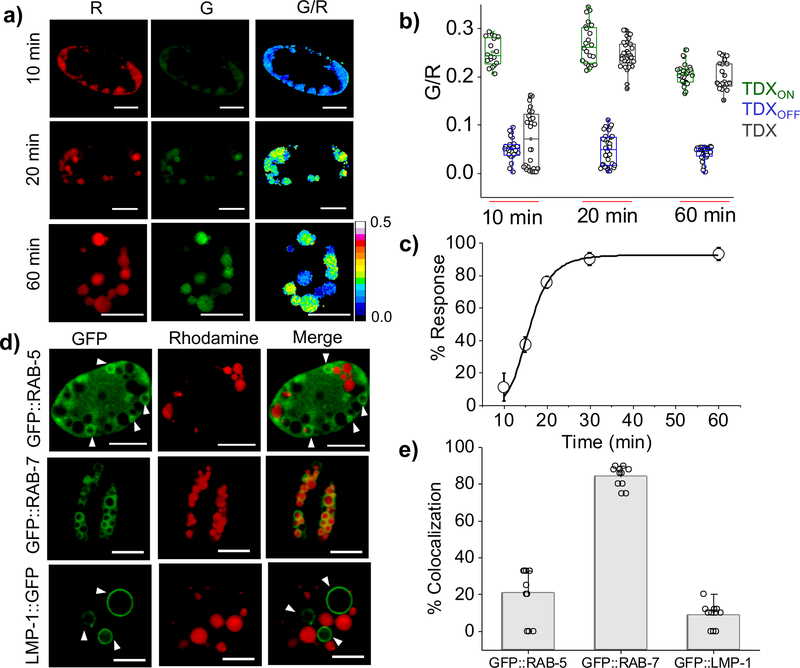
Figure 2: Spatiotemporal detection of thiol disulfide exchange reaction in endo-lysosomal compartment of C. elegans coelomocytes.
a) Representative pseudo-colored images of TDX reporter’s endocytosed by coelomocytes at indicated time points post injection. b) Box plot showing G/R ratio at different time points (n = 20 cells, ≥ 80 endosome). c) Plot of percentage (%) response of TDX reporter at different time point post injection in worms, Error bar indicates the mean of three independent experiments ± s.e.m. d) Colocalization of TDXR with transgenic worms expressing molecular markers of endocytic vesicles such as GFP::RAB-5, GFP::RAB-7 and LMP-1::GFP in coelomocytes, 20 min post injection (n = 10 cells, ≥ 50 endosomes ). Scale bar, 5 μm. e) Quantification of colocalization in (d).
The tripartite reporter comprising TDXON, TDXOFF and TDX, jointly corrects for any pH effects on the fluorescence of the sensor module. For example the lumenal pH decreases along the endocytic pathway which could affect the fluorescein intensity. We correct for this by performing three sets of experiments at each timepoint, injecting either TDXON, TDXOFF or TDX, obtaining G and R images, heat maps and G/R values (Fig. 2b). We calculate the percentage response of TDX (Fig. 2c) as a function of time using the equation 1 (Methods section). This yielded a sigmoidal curve for thiol-disulfide exchange as a function of time where, 20 min post injection the reaction was complete (Fig. 2c).
For the first 10 minutes, TDX response is negligible, and then starts reacting at t = 15 min and by t = 20 minutes, the reaction is ~ 80% complete. Colocalization studies with endosomal markers in transgenic nematodes were performed. Rhodamine labelled dsDNA, denoted TDXR was injected in nematodes expressing GFP::RAB-5 as an early endosomal marker, GFP::RAB-7 as an late endosomal/lysosomal marker and LMP-1::GFP as a lysosomal marker (Fig. 2d). At t = 20 min post injection TDXR showed 85% colocalization with GFP::RAB-7 (Fig. 2e) indicating its localization in the late endosome. This reveals that thiol-disulfide exchange of TDX occurs in late endosomes in coelomocytes.
The observed intra-endosomal disulfide exchange could be mediated by small molecules like cysteine, glutathione, H2S etc., or by enzymes16,27. Using a well-characterized, icosahedral DNA nanocapsule reporter system (SI Fig. 3 & 4)28,29, we found that uncatalyzed thiol-disulfide exchange mediated by small biologically available thiols was negligible.
PDI-3 and TRX-1 catalyse thiol exchange in late endosomes
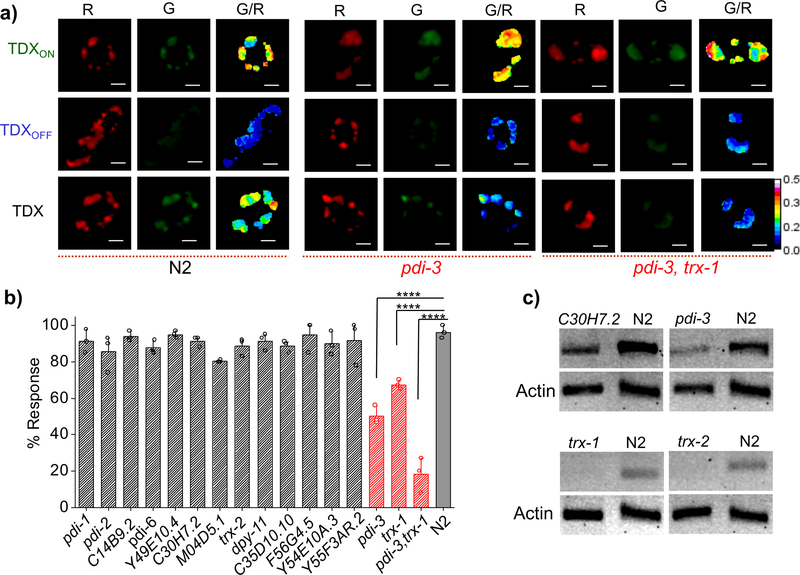
Next, we sought to identify the protein players, if any, that mediated intra-endosomal disulfide reduction in coelomocytes. Proteins that catalyse disulfide exchange generally contain thioredoxin domains e.g., thioredoxins or protein disulfide isomerases30. Using BLASTP31 we found ~23 proteins in the C. elegans genome that contained at least one thioredoxin domain (SI Fig. 16). We excluded seven that contained either a mitochondrial or nuclear localization sequence, which precluded their endosomal localization (SI Table 2). We then knocked down each of the fifteen candidate genes by RNA interference (RNAi) and, in each genetic background, we quantitated the percentage response of the TDX reporter due to intra-endosomal thiol disulfide exchange (Fig. 3b). We obtained two clear hits, where knocking down pdi-3 or trx-1 showed 50% and 40% reduction in disulfide exchange respectively as compared to wild type nematodes (Fig. 3b). Simultaneous knockdown of both pdi-3 and trx-1 in nematodes showed a dramatic reduction of disulfide exchange, nearly comparable to G/R values seen with TDXOFF (Fig. 3a). This indicates that disulfide reduction in the late endosome is predominantly due to pdi-3 and trx-1. The efficiency of RNAi knockdown of relevant genes were confirmed by RT-PCR followed by gel electrophoresis (Fig. 3c).
Figure 3: PDI-3 and TRX-1 are responsible for disulfide exchange reaction inside endocytic vesicles.
a) Representative pseudo-color images of coelomocytes labeled individually with TDXON, TDXOFF and TDX in worms of indicated genetic backgrounds. b) Plot of % response of TDX in different mutants and RNAi knockdowns 20 min post injection. Error bars indicate the mean of three independent experiments ± s.e.m. (n = 10 cells, ≥ 50 endosomes). **** indicates the two tailed p value is less than 0.0001 in unpaired t test, % response of pdi-3, trx-1 and pdi-3, trx-1 double RNAi worms at 20 min post injection. c) RT-PCR analysis of total RNA isolated from wild type (N2) and RNAi worms. PCR amplified cDNA of specified gene products in both RNAi and N2 worms are shown. Actin is used as a loading control. Scale bar, 5 μm.
TRX-1 mainly reduces several disulfide containing proteins, and thereby serves to enhance the activity of stronger reducing proteins e.g., protein disulfide isomerases (PDIs). In fact, the oxidation potential of PDIs are nearly 50 fold higher than thioredoxin32, suggesting that PDI-3 could also function along with TRX-1 at the acidic pH in the late endosome24. Thioredoxin-1 (TRX-1) is maximally present in the cytosol, while PDI-3 is maximally present in the endoplasmic reticulum (ER) due to its strong ER retention signal33. It is notable that despite the presence of PDI-3 and TRX-1 on the plasma membrane, as well as 1–10 μM extracellular glutathione34, our nanodevice reporters are reduced specifically in the late endosome. This reveals the existence of a minor, catalytically active pool of PDI-3 and TRX-1 in the late endosome where they bring about intra-endosomal disulfide exchange.
Endosomal disulfide exchange facilitates pathogen infection
Disulfide exchange is co-opted by pathogens like B. cenocapacia, C. diphtheriae, the HIV virus, the rotavirus ECwt, ganjam virus etc. so that they can efficiently infect host cells35–39. We addressed significance of the catalytic activities of our hits PDI-3 and TRX-1 in endosomes, by investigating their roles in mediating pathogen infection. For this, we used a previously established Corynebacterium diphtheriae infection model in C. elegans39. Briefly, C. diphtheriae secretes an AB toxin called diphtheria toxin (DT), comprising a non-toxic B-subunit (DT-B) and a toxic A-subunit (DT-A) linked through a disulfide bond. This disulfide bond is reduced within the endosomes and DT-A thereby translocates into the cytosol, inhibiting protein synthesis in the host cell. Disulfide reduction is thus a critical step to bring about toxin infection (SI. Fig. 8).
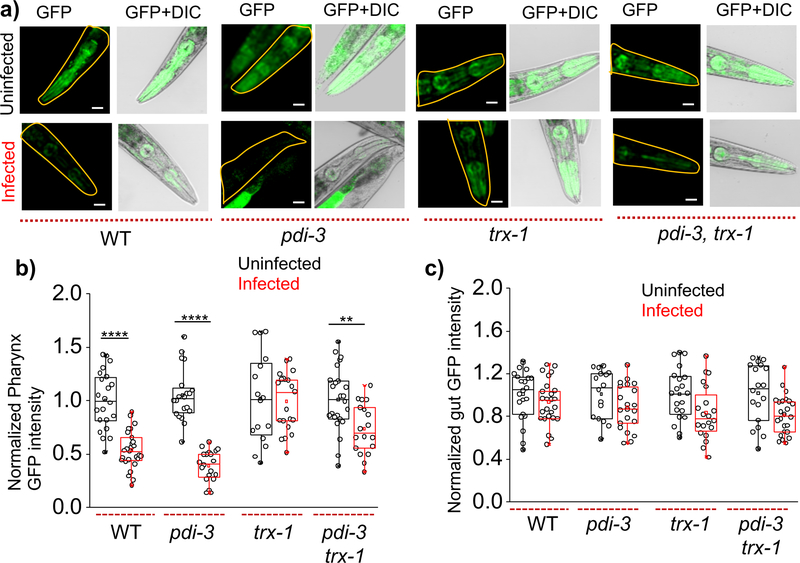
We used the transgenic nematode cl2070, which expresses GFP under a heat shock promoter hsp-16.2 in all tissues. Successful infection by C. diphtheria is expected to result in cytosolic DT-A compromising protein synthesis leading to nematodes that fail to express GFP in the pharynx upon heat-shock40. Accordingly, nematodes infected with C. diphtheriae showed significantly less GFP in the pharynx compared to non-infected nematodes (Fig. 4a). Notably, GFP intensity in the gut were comparable in both infected and non-infected nematodes as the site of action of C. diphtheriae is not the gut, but the pharynx (Fig. 4b–4c and SI Fig. 6–7)39.
Figure 4: Thioredoxin-1 offers protection against DT infection.
a) Representative pseudo-color and overlaid images of pharyngeal GFP expression in indicated genetic backgrounds of nematodes infected or not infected with C. diphtheriae. Box plots of normalized b) pharyngeal and c) intestinal GFP intensity of infected and non-infected nematodes with indicated genetic background (n = 20 worms). GFP intensity distribution is normalized by mean intensity of respective not infected worm. ****p <0.0001 and **p < 0.005 obtained using unpaired t-test. Scale bar, 100 μm. Experiments are replicated in duplicates.
Next, we examined the consequences of inhibiting intra-endosomal disulfide reduction on DT infection, by knocking down pdi-3 and/or trx-1 in this infection model. We observed that trx-1, but not pdi-3, knockdowns show retention of pharyngeal GFP expression upon treatment with C. diphtheriae (Fig. 4a–4b). This indicates that by knocking down endosomal disulfide exchange brought about by the minor endosomal population of thioredoxin-1 impedes DT infection by reducing the extent of cytosolic translocation of the DT-A. Knocking down both pdi-3 and trx-1 also prevents DT infection (Fig. 4b). In coelomocytes, we observe that PDI-3 and TRX-1 both are involved in endosomal disulfide reduction but in the case of DT infection only TRX-1 is responsible. Even though both TRX-1 and PDI-3 were responsible for intra-endosomal disulfide reduction, we found that DT infection depends on the minor pool of TRX-1, and not PDI-3. This is also consistent with in vitro studies that show that the intra-chain disulfide bond of DT is a good substrate for thioredoxin-1 at acidic pH41. Our findings imply that DT preferentially exploits intra-endosomal reduction by thioredoxin-1 in vivo and that inhibiting this minor pool could be a promising route to impede DT infection.
TRX-1 mediates endosomal disulfide exchange in human cells
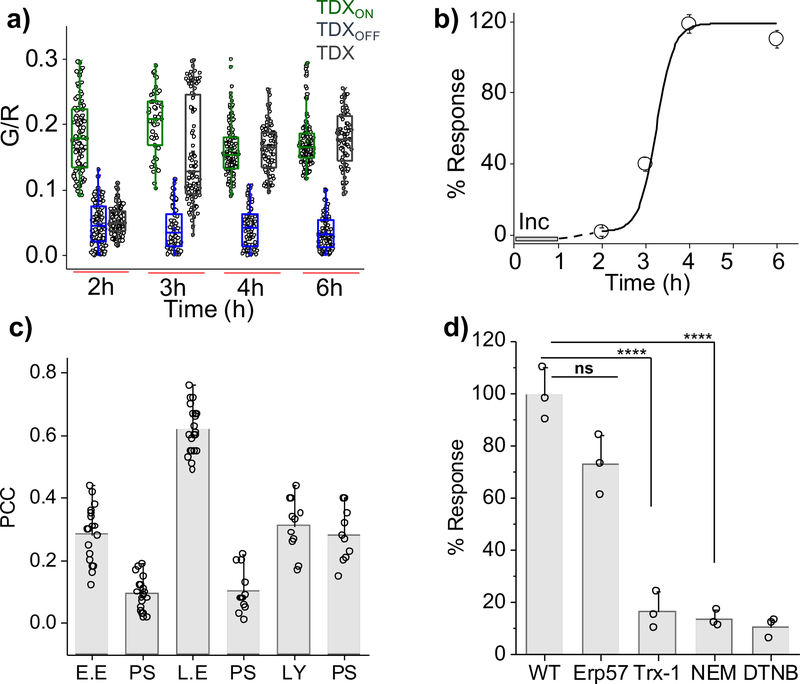
Next, we sought to generalize this reporter technology by probing disulfide reduction in a mammalian cell culture system. All TDX reporters undergo scavenger receptor mediated endocytosis in HeLa cells upon incubation with 1 μM of any TDX reporter42. We could therefore follow disulfide reduction as a function of time using TDX reporters as described (SI Fig.11). While we observed no disulfide reduction for the first 2 h, TDX reporters started responding at t = 3 h and the reaction was complete by t = 4 h (Fig. 5a–5b). The analogous experiment with TDXOFF showed negligible change in G/R ratio (Fig. 5a). We pharmacologically inhibited disulfide isomerases in HeLa cells using N-ethyl maleimide (NEM) or by blocking surface and endosomal thiols using 5,5’-dithio-bis-(2-nitrobenzoic acid) (DTNB). Both treatments reduced reporter response by ~90% confirming that endosomal disulfide reduction was indeed responsible for the signal increase (Fig. 5d & SI Fig. 12).
Figure 5: TRX-1 mediates endosomal disulfide reduction in mammalian cells.
a) Box plot of G/R values of the TDX reporter system in HeLa cells at indicated time points post internalization (n = 20 cells, ≥150 endosomes). b) Percentage (%) response of the TDX reporter system as a function of time, [Inc. = incubation period, line serves as a visual aid]. c) Pearson’s correlation coefficient (PCC) of colocalization between pulsed TDXA647 after a 3 h chase in HeLa cells expressing endosomal markers. [E.E. = early endosome, Rab5-RFP, L.E. = late endosome, Rab7-RFP), Ly = lysosome, TMR dextran, PS = pixel shift] (n ≥ 10 cells), d) Percentage (%) response of the TDX reporter system at t = 4 h in HeLa cells treated with indicated inhibitors or siRNAs against indicated proteins, [NEM = N-ethyl maleimide, DTNB = 5, 5-dithio-bis-(2-nitrobenzoic acid)]. Error bar corresponds to s.e.m. from three independent experiment, ns = not significant, ****p <0.0001 obtained using unpaired t-test.
Colocalization studies with various endosomal markers indicated that the observed disulfide reduction occurred in the late endosome. We pulsed HeLa cells expressing Rab5-RFP as an early endosomal marker or Rab7-RFP as a late endosomal marker TDX bearing a single Alexa-647 fluorophore (TDXA647) and assayed for colocalization after a 3 h chase. We observed ~70% colocalization with Rab7-RFP and insignificant colocalization with Rab5-GFP. Lysosomes were labeled with TMR-dextran through fluid phase endocytosis after a 1 h pulse and 12 h chase. HeLa cells pre-labelled thus were then pulsed and chased with TDXA647 as above showed negligible colocalization with lysosomes (Fig. 5c).
To identify the major protein player involved in this reduction, we knocked down the human analogues of PDI-3 and TRX-1 by RNAi. HeLa cells where Erp57 and TRX-1 were each knocked down were investigated using the tripartite reporter system and the G/R ratios were obtained. We observed that disulfide reduction in TRX-1 depleted cells was reduced by >70% compared to normal cells, while Erp57 depleted cells showed negligible change (Fig. 5d, SI Fig. 12). This indicates that in mammalian cells, a minor pool of endosomal resident TRX-1 plays a major role in endosomal disulfide reduction.
Summary
The modular design of our DNA-based tripartite fluorescent reporter system enables quantitative imaging of intra-endosomal disulfide exchange in situ in coelomocytes of C. elegans as well as in mammalian cells. It revealed that thiol-disulfide exchange along the endolysosomal pathway occurs mainly in the late endosome. We ruled out small molecule thiol-mediated reduction in late endosomes, implicating the action of proteins in these acidic compartments. In fact, the pKa of the N-terminal active site cysteine of PDI can be as low as 4.8, while that of thioredoxins can range between ~6.0–7.043. This is in consistent with the rapid thiol reduction kinetics we observe in the acidic late endosome lumen, supporting that disulfide exchange is likely to be enzyme catalyzed.
Quantitative imaging of thiol-disulfide exchange in RNAi knockdowns of various candidate genes revealed that PDI-3 and TRX-1 were the enzymes responsible for the observed intra-endosomal disulfide reduction in vivo. Though PDI-3 and TRX-1 are predominantly localized in the endoplasmic reticulum (ER) and cytosol respectively33,44 a minor pool of these proteins is localized in sub-cellular compartments where they perform critical functions. Using a diphtheria toxin infection model, we showed that inhibiting the catalytic activity of this minor population of enzyme inside the endosome protects the host cell from pathogen infection. The reporter system revealed that TRX-1 was also responsible for disulfide reduction in late endosomes of human cells. Given the large number of toxins and pathogens that exploit intra-endosomal disulfide reduction to bring about infection38, our work suggests that the selective inhibition of intra-endosomal disulfide exchange could be of interest to develop anti-infectives.
The advantage of this tripartite reporter system is the newfound ability to directly and selectively assay, in live cells and with spatial information, the enzyme activity of protein populations in organelles by localizing the detection chemistry within the organelle (SI Figure 14–15). The stoichiometric precision of DNA hybridization enables two major advantages, unique to DNA-based probes and inaccessible to any other reporter technology: (i) molecularly identical probes, with fixed, uniform sensor to normalizer ratios that quantify a given analyte with accuracies unmatched by any other molecular scaffold. (ii) “Plug-and-play” capability due to its modular nature: Once a DNA-probe is validated, it can be directly used in a host of other organelle locations by simply swapping out one organelle targeting module for any other.
It can thus be readily integrated with existing diversity of small molecule reporters and enzyme-cleavage chemistries to provide robust readouts exclusively in organelles in situ within living cells (SI table 3). For example, lysosomal enzyme chemistries may be integrated to measure lysosomal enzyme activity in situ with several applications in lysosomal storage disorders45. Catalytic activity of endosomal proteases such as cathepsin B, cathepsin D, or β-secretase-1 results in protein processing events leading to cancer and Alzheimer’s disease46,47. Protease action by proprotein convertases in the Golgi underlie neurogenesis, TGF-β signalling and AB5 toxin infection48. The ability of DNA nanodevices to selectively quantitate sub-cellular enzymatic cleavage in live cells could provide new diagnostics and drug discovery assays.
It can be also be integrated with newer reporting strategies that probe enzymatic function such as activity based protein probes to identify enzymatic activity in cellular compartments49. The advent of transformative technologies such as fluorescent proteins, the SNAP-tag50 or the Halo-tag57 ushered in a new era of visualizing proteins56 in situ by enabling methods to image the major population of any protein of interest. The organelle-targetable nanodevice system described here effectively complements these powerful technologies by selectively interrogating minor populations of enzymes involved critical cellular functions, which cannot otherwise be studied.
Methods
Materials and Methods.
All the chemicals used for the synthesis were purchased from commercial source (Sigma, USA). 1H NMR and 13C NMR spectra of the newly synthesized compounds were recorded on a Bruker AVANCE II+, 500MHz NMR spectrophotometer. TMS is used as an internal standard. Mass spectra were recorded in Agilent 6224 Accurate-Mass TOF LC/MS. HPLC purified oligonucleotides conjugated with either fluorophore or azide functional group were obtained from Integrated DNA Technologies (IDT, USA). All oligonucleotides are ethanol precipitated and quantified by UV absorbance at λ= 260 nm.
Synthesis of TDX reporter.
Rhodamine and azide labelled DNA-strand (O-Rhodamine Red X and O-azide) (Table-1) were mixed in equimolar concentration (25 μM each) in 20 mM phosphate buffer containing 100 mM KCl at pH 7. The mixture was heated at 90°C for 10 min and then cooled down to room temperature at the rate of 5°C/15 min and stored in 4°C for overnight to form a complete DNA-duplex.
Next, Rhodamine DNA-duplex was conjugated with compound 8 by reported copper catalysed azide-alkyne click chemistry protocol51. Initially 20 μL rhodamine conjugated DNA duplex was (25 μM) diluted in 13.5 μL water, to that of 3 μL compound 8 (5 mM) in DMSO was added. Then 1:1 (v/v) premix solution of 7.5 μL of CuSO4 (0.1 M) and THPTA (0.2 M) was added to it. The reaction mixture was degassed with N2 for 2 min and followed by this 6 μL of sodium ascorbate (0.1 M) was added. The mixture was further degassed for 0.5 min and then stirred at room temperature for 1 h under N2 atmosphere. Native PAGE (20 wt%) showed complete formation of TDX reporter (Fig. 1a and Supplementary Fig. 1a). Subsequently, the reaction mixture was diluted with pH = 6.0 phosphate buffer and washed with 10% acetonitrile (to remove excess of compound 8 using Amicon Ultra-0.5mL centrifugal filter (MWCO 3 kDa). The same washing procedure was continued until the filtrate showed no trace of compound 8 using fluorescence spectroscopy (λem = 520 nm). Followed by, we performed native gel electrophoresis and monitored by fluorescence which reveals negligible single strand in our preparations.
Synthesis of TDXON reporter.
10 μL TDX reporter (8 μM) was mixed with 1 μL (5 mM) NaSH solution in 0.1 M phosphate buffer at pH 7.4 and stirred for 2 h at room temperature. Followed by this, excess thiol was removed by ultracentrifugation using Amicon filter (MWCO 3 kDa) to obtain TDXON reporter (see Fig. 1a).
Synthesis of TDXOFF reporter.
TDXOFF reporter was prepared in a similar procedure like TDX reporter, except compound 9 was used here instead of compound 8. The formation of TDXOFF reporter was characterized by using gel electrophoresis (see Supplementary Fig. 1a).
Conjugation of compound 8 with azido dextran.
2 mg Azido functionalized dextran (10 kDa, 2–3 azido group per dextran) was dissolved in 90 μL milli Q water. 20 μL of compound 8 (5mM), followed by 50 μL CuSO4 (0.1 M) and THPTA (0.2 M) mixture (1:1) was added to it. The reaction mixture was degassed for 2 min and 40 μL sodium ascorbate (0.1 M) was added, purged again with N2 for 0.5 min before stirring at room temperature for 1 h under N2 atmosphere. The resulting dextran conjugate was diluted with milli Q water and washed with the 10% acetonitrile to remove excess of compound 8 using an Amicon filter (MWCO 3 kDa).
Self-assembly of IA647FD.
Half icosahedrons (VU5 and VL5) was prepared as describe in literature52. In order to synthesize the DNA-icosahedron, VU5 and VL5 (3 μM each, 30 μL, in 50 mM phosphate buffer, pH 6.0) containing 2 mM solution of the compound 8 conjugated dextran (FD) (M. Wt.~10 kDa) was mixed in an eppendorf tube and heated to 37 °C for 30 min. The temperature was brought down to 20 °C with a rate of 1 °C/ 3 min and followed by incubation for 2 h. The reaction mixture was transferred to refrigerator at 4°C for further incubation for longer time periods up to 48 h. The formation of IA647FD was characterized by 0.8 wt% Agarose gel electrophoresis (Supplementary Fig. 3a).
Preparation of IA647FD-ON.
10 μL solution (3 μM) of IA647FD was treated with 1 μL (5 mM) NaSH solution in 0.1 M phosphate buffer at pH 7.4 and stirred for 2 h at room temperature. The excess thiol was removed by ultracentrifugation using Amicon filter (MWCO 3 kDa).
Determination of size of different thiol.
DLS experiment was done in Wyatt Dynapro Nanostar & Plate Reader. Samples were dissolved in milli-Q water or buffer filtered through 0.22 μm filter to remove dust particles. Samples were illuminated with laser wavelength of 658 nm, at a sensitivity of 80% and a scattering angle set at 90° with a 10 second acquisition time for data collection. Percentage intensity observed for each sample, was plotted against respective Rh values. The size of PEG-SH (3.2 kDa), Dex-SH (10 kDa) and Dex-SH (40 kDa) was determined from DLS measurements. Aqueous solution of the free thiol containing polymer (1 mg/mL) was again filtered in a dust-free environment, and DLS measurement was carried out with this solution. For glutathione, cysteine and H2S, we used Chem 3D ultra 8.0 software and used the end to end distance of the energy minimized structure as diameter of those molecules.
In vitro fluorescence measurements.
Fluorescence spectra were recorded on a FluoroMax-4 instrument (Horiba Jobin Yvon). TDX and TDXOFF reporter was diluted to 100 nM in 0.1 M phosphate buffer at pH = 7.2 in presence or absence of 5 mM GSH. These samples were excited at 450 nm (for fluorescein emission) and 575 nm (for rhodamine emission). The emission spectrum was collected between 460–600 nm and 590–700 nm for fluorescein and rhodamine respectively at different time point. Three independent measurements were recorded for each sample (Fig. 1c). We checked the specificity of reporter dye towards thiol disulfide exchange reaction; we treated the reporter dye (compound 8) with different metal ions and amino acids. We have observed no reaction with any of the metal ions and amino acids tested except cysteine (Supplementary Fig. 1c).
In order to check the substrate availability for disulfide exchange reaction with IA647FD as a function of the size of reactive thiols (1 mM), we have treated IA647FD (3 μM) and FD with various thiol and checked the emission spectra after 1 h of incubation. For monitoring the emission spectra, the samples were excited at 450 nm (fluorescein channel) and 647 nm (Atto 647N channel). The emission spectrum was collected between 460–600 nm and 650–750 nm for fluorescein and Atto 647N respectively. (Supplementary Fig. 3b).
Protocols for C. elegans and strains.
Standard methods for the maintenance of C. elegans were followed. We used wild type C. elegans strain isolated from Bristol (Strain N2). Mutant strain used for the experiment is VC586 [pdi-1(gk271) III].
Transgenic strains used for this study are;
cdIs131 [pcc1::GFP::rab-5 + unc-119(+) + myo-2p::GFP], a transgenic strain that express GFP-fused early endosomal marker RAB-5 inside coelomocytes.
cdIs66 [pcc1::GFP::rab-7 + unc-119(+) + myo-2p::GFP], a transgenic strain that express GFP-fused late endosomal / lysosomal marker RAB-7 inside coelomocytes.
pwIs50 [lmp-1::GFP + Cbr-unc-119(+)], a transgenic strain that express GFP-fused lysosomal marker LMP-1.
dvIs70 [hsp-16.2p::GFP + rol-6(su1006)], a transgenic strain which have GFP expression under heat shock protein HSP-16.2 promoter. It express GFP in presence of mild heat shock.
Protocols for C. diphtheria strains and growth conditions.
Two different kind of C. diphtheria strain have been used for pathogen infection to C. elegans. They are toxic C. diphtheria (ATCC 13812) which produces diphtheria toxin (DT) and less toxic C. diphtheria (ATCC 39526) (DTΔ149) which produces mutated diphtheria toxin. For toxic and less toxic C. diphtheria, we have used tryptic soy broth with 5 % sheep blood and ATCC medium: 1417 Low Iron YC medium as a culture medium respectively and inoculate for 24 h at 37°C as described in ATCC website.
TDX and IA647FD reporters targeted to coelomocyte of C. elegans.
For coelomocyte targeting52, we microinjected 2 μM of TDX reporter in the dorsal side in the pseudocoelom, just opposite to the vulva, of 1-day-old wild type hermaphrodites. Injected worms then placed in a new NGM agar containing petriplate at 22°C for incubation at different time point. Followed by this they were mounted in an Agar pad (2.0%) and anaesthetized using 40 mM sodium azide in M9 buffer and performed fluorescence imaging experiment. Same protocol was followed in the case of TDXOFF and TDXON.
DNA-icosahedron encapsulated reporters, (IA647FD, IA647FD-ON and IA647FD-OFF, 3 μM each) were injected to the wild type worms as described earlier and imaged at 30 min post-injection period.
Colocalization of TDXR with late endosomes in coelomocytes.
In order to investigate the localization of TDX reporter at 20 min post injection, we have conducted co-localization experiment using cdIs131, cdIs66 and pwIs50 transgenic worms. For this, we have injected 2 μM rhodamine DNA-duplex to these worms and imaged after 20 min. We have observed ~ 85% co-localization of TDX reporter with RAB-7::GFP positive vesicles, which marks late endosomal and lysosomal compartments (see Fig. 2d and Supplementary Fig. 8).
RNAi experiment.
We used BLASTP53 to look for thioredoxin domain containing protein in C. elegans genome. Bacteria of our interest, expressing double-stranded RNA (see Supplementary table 2) were obtained from Ahringer RNAi library54 and Vidal Unique55. Cloned bacteria were fed to the worms and ~ 60 one-day adults of the F1 progeny were used for screening. The Ahringer or Vidal unique library did not contain bacterial clones for Y73B6BL.2 and hence this gene was not included in this screen.
We assayed mRNA levels of the candidate genes by RT – PCR. Briefly, we isolated total RNA using the Trizol-chloroform method; 2.5 μg of total RNA was converted to cDNA using oligo-dT primers. 5 μl of the reverse transcription reaction was used to set up a PCR using gene-specific primers. PCR products were analysed on a 1.5 % agarose-TAE gel (see Fig. 3c). Size of the PCR products expected for each gene were: actin (360 bp), pdi-3 (682 bp), C30H7.2 (798 bp), trx-2 (316 bp), trx-1 (261 bp).
hsp-16.2::GFP experiments to measure DT infection-triggered translational inhibition.
hsp2::GFP experiments were modified from literature reported procedure56. Briefly the fresh culture of either toxic or less toxic C. diphtheria (DTA149) were seeded into the nematode growth medium (NGM) containing agar plate. Each plates contains 50 μL of bacterial culture. After that the plates were kept at 25°C for 24 h for drying the liquid medium on agar. WT or different genetic background worms were washed onto C. diphtheria seeded plates and incubated for 6 h for infection at 25°C followed by washed and transferred to a plat containing 0P50 or the bacteria carrying the RNAi clone in it. These C. elegans then subjected to 1 h heat shock at 37°C and then returned to 25°C for another 1 h. After that the worms are imaged for GFP expression. GFP expression was quantified by keeping the microscope settings fixed for all samples. The standard region of pharynx and intestine was chosen for GFP intensity measurement (see Supplementary Fig. 6). For the intestine we chose a region towards the posterior of intestine because it showed the robust GFP expression. The GFP intensity was measured for at least 20 worms in each genetic background worm.
Cell culture.
HeLa cells were a kind gift from Prof. Chuan He (Department of Chemistry, the University of Chicago). Mouse macrophages J774A.1cells were a kind gift from Prof Deborah Nelson, (Department of Pharmacological and Physiological Sciences, the University of Chicago). These cells were cultured in DMEM-F12 with 10% FBS, 100 U/mL penicillin, 100 μg/ mL of streptomycin and maintained at 37 oC under 5% CO2. Bone marrow derived macrophages (BMDMs) were isolated from femurs and tibia of male C57BL/6 mice and were obtained from Prof. Lev Becker lab (Ben May Department for Cancer Research, University of Chicago). Cells were differentiated to macrophages using murine M-CSF for 6 days. For LPS activation, cells were treated with 20 ng/mL LPS for 24 h. All animal studies are in compliances with ethical regulations approved by the University of Chicago Institutional Animal Care and Use Committee (ACUP 72209).
Microscopy and ratiometric image analysis.
Wide-field microscopy was performed on an IX83 inverted microscope (Olympus Corporation of the Americas, Center Valley, PA, USA) using a 60X, 1.42 NA, phase contrast oil immersion objective (PLAPON, Olympus Corporation of the Americas, Center Valley, PA, USA) and Evolve® Delta 512 EMCCD camera (Photometrics, USA). Filter wheel, shutter and CCD camera were controlled using Metamorph Premier Version 7.8.12.0 (Molecular Devices, LLC, USA), appropriate for the fluorophore used. Images on the same day were acquired under the same acquisition settings. Confocal imaging was carried out on a Leica SP5 II STED-CW super-resolution microscope, using an Argon ion laser for 488 nm excitation and DPSS for 561 nm excitation with a set of dichroic, excitation, and emission filters suitable for each fluorophore. Crosstalk and bleed-through were measured and found to be negligible between GFP / fluorescein and Rhodamine.
All the images were background subtracted by using mean intensity calculated from an adjacent cell-free area. Fluorescein and Rhodamine images were colocalized and endosomes showing good colocalization were analysed using ImageJ-Win64 software (NIH). Mean fluorescence intensity in each endosome was measured in fluorescein (G) and rhodamine (R) channels. A ratio of G/R intensities was calculated from these values. Pseudocolour images were generated by measuring the G/R ratio per pixel. Using ImageJ software, pixels were then colour coded accordingly to indicate differences between high and low G/R ratios.
% Response Calculation.
Percentage response of the TDX reporter at different time points for the wild type worm and for the RNAi worm (20 min) were calculated using the following equation (1).
| (1) |
(Here, G/RTDX, G/RTDXOFF and G/RTDXON indicate the observed fluorescence intensity ratio of fluorescein to that of rhodamine at a given time point by using TDX, TDXON and TDXOFF reporters respectively).
Targeting TDX reporter to endolysosomal pathway in HeLa cells.
We incubated HeLa cells with TDX reporter (1 μM) containing DMEM without FBS for 1 h at 37 °C. Cells were then washed with PBS (pH=7.4) thrice to remove any un-internalized TDX nanodevice and incubated for various chase times in DMEM complete media. Prior to imaging, cells were washed with PBS and imaged in HBSS (Hank’s Balanced Salt Solution, GE healthcare) buffer. Similar protocol was followed for TDXON and TDXOFF reporters.
RNAi knockdown and pharmacological inhibition in HeLa cells.
All relevant siRNAs were procured from Dharmacon, Inc. HeLa cells were split on imaging dish at 1×104 seeding density. After 16 h in culture, the cells were transfected with siRNAs using DharmaFECT1 (Dharmacon; Catalogue No: T-2001–01) transfection reagent according to the manufacturer’s protocol. For Erp57 and TRX-1 knockdown, HeLa cells were transfected with Erp57 siRNA57 (5’-GGGCAAGGACUUACUUAUUTT-3 ‘) and TRX-1 siRNA (Dharmacon, Inc., Catalogue No: M-006340–010005). As a negative control, we used the NC siRNA (Dharmacon, Inc., Catalogue No: D-001210–01-05). After 72 h of transfection, cells were incubated with the tripartite TDX reporter for 1 h in DMEM without FBS and chased for 3 h in complete media and imaged as described earlier.
For pharmacological inhibition, cells were incubated with complete media containing either NEM (10 μM) or DTNB (100 μM) for 1 h, followed by 1 h of incubation with TDX reporter in DMEM without FBS. The cells were chased for 4 h in complete media containing either 10 μM NEM or 100 μM DTNB, washed with PBS and imaged as described in HBSS buffer containing either 10 μM NEM or 100 μM DTNB so that the relevant pharmacological inhibitor was always present.
Cathepsin C activity assay.
J774A.1 cells with or without Cathepsin inhibitor E64 were pulsed with 500 nM of either Cat or CatON probes (SI Figs. 14 & 15) separately for 30 min at 37° C. After 1 h, cells were washed and imaged in the Rhodamine (G, λem = 520 nm) and Alexa 647 (R, λem = 670 nm) channels. The CatON probe provides the maximum possible ratiometric signal (G/R) if the Cat probe was completely cleaved. All G/R values were normalized to that of CatON. Similar pulse and chase experiments were performed in BMDMs with 200 nM Cat probes. BMDMs were then subjected to flow cytometry on a FACSCanto™ II flow cytometer (SI Fig.15).
Statistical Analysis.
We used Graphpad unpaired t-test calculator for all statistical significance calculations (https://www.graphpad.com/quickcalcs/ttest1/).
Supplementary Material
Acknowledgments
We thank Jon Clardy, John Kuriyan, Souvik Modi and Ai Lin Chun for valuable comments on this work. We thank the Integrated Light Microscopy facility at the University of Chicago, the Caenorhabditis Genetic Center (CGC), Johnny Fares and Mark Edgley for C. elegans strains, Michael Glotzer and Frederick M. Ausubel for RNAi clones and valuable discussions, Sean Crosson for BSL facility for C. diphtheriae work, Chang Cui for bone marrow derived macrophages and flow cytometry. This work was supported by the University of Chicago Women’s Board, Chicago Biomedical Consortium with support from the Searle Funds at The Chicago Community Trust, C-084 as well as a CBC post-doctoral research grant (PDR-073) and University of Chicago start-up funds to Y.K. Y.K. is a Brain Research Foundation Fellow.
Footnotes
Competing financial interest. The authors do not declare any competing financial interest.
Additional Information. Supplementary information is available in the online version of the paper.
Data Availability Statement:
The data that support the plots within this paper and other finding of this study are available from the corresponding author upon reasonable request.
Reporting Summary. Further information on research design is available in the Nature Research Reporting Summary linked to this article.
References
- 1.Huang S-H, Li Y, Zhang J, Rong J & Ye S Epidermal growth factor receptor-containing exosomes induce tumor-specific regulatory T cells. Cancer Invest 31, 330–335 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Efeyan A, Comb WC & Sabatini DM Nutrient-sensing mechanisms and pathways. Nature 517, 302–310 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dean N & Pelham HR Recycling of proteins from the Golgi compartment to the ER in yeast. J. Cell Biol. 111, 369–377 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhuniya S et al. An activatable theranostic for targeted cancer therapy and imaging. Angew. Chem. Int. Ed. Engl. 53, 4469–4474 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Lee MH et al. Hepatocyte-targeting single galactose-appended naphthalimide: a tool for intracellular thiol imaging in vivo. J. Am. Chem. Soc. 134, 1316–1322 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Crivat G & Taraska JW Imaging proteins inside cells with fluorescent tags. Trends Biotechnol. 30, 8–16 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Los GV et al. HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem. Biol. 3, 373–382 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Gething MJ & Sambrook J Protein folding in the cell. Nature 355, 33–45 (1992). [DOI] [PubMed] [Google Scholar]
- 9.Mesecke N et al. A disulfide relay system in the intermembrane space of mitochondria that mediates protein import. Cell 121, 1059–1069 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Burgoyne JR et al. Cysteine redox sensor in PKGIa enables oxidant-induced activation. Science (80-.). 317, 1393–1397 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Wang M-Y et al. A redox switch in C-reactive protein modulates activation of endothelial cells. FASEB J. 25, 3186–3196 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Collins DS, Unanue ER & Harding CV Reduction of disulfide bonds within lysosomes is a key step in antigen processing. J. Immunol. 147, 4054–4059 (1991). [PubMed] [Google Scholar]
- 13.Guermonprez P et al. ER-phagosome fusion defines an MHC class I cross-presentation compartment in dendritic cells. Nature 425, 397–402 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Stolf BS et al. Protein disulfide isomerase and host-pathogen interaction. ScientificWorldJournal 11, 1749–1761 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maiti S et al. Gemcitabine-coumarin-biotin conjugates: a target specific theranostic anticancer prodrug. J. Am. Chem. Soc. 135, 4567–4572 (2013). [DOI] [PubMed] [Google Scholar]
- 16.FeenerS E, Shene W-C & Ryser H Cleavage of Disulfide Bonds in Endocytosed Macromolecules. [PubMed] [Google Scholar]
- 17.Shen WC, Ryser HJ & LaManna L Disulfide spacer between methotrexate and poly(D-lysine). A probe for exploring the reductive process in endocytosis. J. Biol. Chem. 260, 10905–10908 (1985). [PubMed] [Google Scholar]
- 18.Meyer AJ & Dick TP Fluorescent protein-based redox probes. Antioxid. Redox Signal. 13, 621–650 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Chakraborty K, Veetil AT, Jaffrey SR & Krishnan Y Nucleic Acid-Based Nanodevices in Biological Imaging. Annu. Rev. Biochem. 85, 349–373 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J, Cao Z & Lu Y Functional nucleic acid sensors. Chem. Rev. 109, 1948–1998 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Famulok M, Hartig JS & Mayer G Functional aptamers and aptazymes in biotechnology, diagnostics, and therapy. Chem. Rev. 107, 3715–3743 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Modi S, Nizak C, Surana S, Halder S & Krishnan Y Two DNA nanomachines map pH changes along intersecting endocytic pathways inside the same cell. Nat. Nanotechnol. 8, 459–467 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Saha S, Prakash V, Halder S, Chakraborty K & Krishnan Y A pH-independent DNA nanodevice for quantifying chloride transport in organelles of living cells. Nat. Nanotechnol. 10, 645–651 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Surana S, Bhat JM, Koushika SP & Krishnan Y An autonomous DNA nanomachine maps spatiotemporal pH changes in a multicellular living organism. Nat Commun 2, 340 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Gutscher M et al. Real-time imaging of the intracellular glutathione redox potential. Nat. Methods 5, 553–559 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Romero-Aristizabal C, Marks DS, Fontana W & Apfeld J Regulated spatial organization and sensitivity of cytosolic protein oxidation in Caenorhabditis elegans. Nat Commun 5, 5020 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang J, Chen H, Vlahov IR, Cheng J-X & Low PS Evaluation of disulfide reduction during receptor-mediated endocytosis by using FRET imaging. Proc. Natl. Acad. Sci. USA 103, 13872–13877 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhatia D et al. Icosahedral DNA nanocapsules by modular assembly. Angew. Chem. Int. Ed. Engl. 48, 4134–4137 (2009). [DOI] [PubMed] [Google Scholar]
- 29.Veetil AT, Jani MS & Krishnan Y Chemical control over membrane-initiated steroid signaling with a DNA nanocapsule. Proc. Natl. Acad. Sci. USA 115, 9432–9437 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sevier CS & Kaiser CA Formation and transfer of disulphide bonds in living cells. Nat. Rev. Mol. Cell Biol. 3, 836–847 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Altschul SF, Gish W, Miller W, Myers EW & Lipman DJ Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 32.Hawkins HC, Blackburn EC & Freedman RB Comparison of the activities of protein disulphide-isomerase and thioredoxin in catalysing disulphide isomerization in a protein substrate. Biochem. J. 275 ( Pt 2), 349–353 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eschenlauer SCP & Page AP The Caenorhabditis elegans ERp60 homolog protein disulfide isomerase-3 has disulfide isomerase and transglutaminase-like cross-linking activity and is involved in the maintenance of body morphology. J. Biol. Chem. 278, 4227–4237 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Smith CV, Jones DP, Guenthner TM, Lash LH & Lauterburg BH Compartmentation of glutathione: implications for the study of toxicity and disease. Toxicol. Appl. Pharmacol. 140, 1–12 (1996). [DOI] [PubMed] [Google Scholar]
- 35.Rubartelli A, Bajetto A, Allavena G, Wollman E & Sitia R Secretion of thioredoxin by normal and neoplastic cells through a leaderless secretory pathway. J. Biol. Chem. 267, 24161–24164 (1992). [PubMed] [Google Scholar]
- 36.Dihazi H et al. Secretion of ERP57 is important for extracellular matrix accumulation and progression of renal fibrosis, and is an early sign of disease onset. J. Cell Sci. 126, 3649–3663 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Pacello F, D’Orazio M & Battistoni A An ERp57-mediated disulphide exchange promotes the interaction between Burkholderia cenocepacia and epithelial respiratory cells. Sci. Rep. 6, 21140 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lasecka L & Baron MD The nairovirus nairobi sheep disease virus/ganjam virus induces the translocation of protein disulphide isomerase-like oxidoreductases from the endoplasmic reticulum to the cell surface and the extracellular space. PLoS One 9, e94656 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ott L et al. Evaluation of invertebrate infection models for pathogenic corynebacteria. FEMS Immunol. Med. Microbiol. 65, 413–421 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Dunbar TL, Yan Z, Balla KM, Smelkinson MG & Troemel ERC elegans detects pathogen-induced translational inhibition to activate immune signaling. Cell Host Microbe 11, 375–386 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moskaug JO, Sandvig K & Olsnes S Cell-mediated reduction of the interfragment disulfide in nicked diphtheria toxin. A new system to study toxin entry at low pH. J. Biol. Chem. 262, 10339–10345 (1987). [PubMed] [Google Scholar]
- 42.Patel PC et al. Scavenger receptors mediate cellular uptake of polyvalent oligonucleotide-functionalized gold nanoparticles. Bioconjug Chem. 21, 2250–2256 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karala A-R, Lappi A-K & Ruddock LW Modulation of an active-site cysteine pKa allows PDI to act as a catalyst of both disulfide bond formation and isomerization. J. Mol. Biol. 396, 883–892 (2010). [DOI] [PubMed] [Google Scholar]
- 44.Wu C et al. Thioredoxin 1-mediated post-translational modifications: reduction, transnitrosylation, denitrosylation, and related proteomics methodologies. Antioxid. Redox Signal. 15, 2565–2604 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Futerman AH & van Meer G The cell biology of lysosomal storage disorders. Nat. Rev. Mol. Cell Biol. 5, 554–565 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Olson OC & Joyce JA Cysteine cathepsin proteases: regulators of cancer progression and therapeutic response. Nat. Rev. Cancer 15, 712–729 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Vassar R, Kovacs DM, Yan R & Wong PC The beta-secretase enzyme BACE in health and Alzheimer’s disease: regulation, cell biology, function, and therapeutic potential. J. Neurosci. 29, 12787–12794 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seidah NG & Prat A The biology and therapeutic targeting of the proprotein convertases. Nat. Rev. Drug Discov. 11, 367–383 (2012). [DOI] [PubMed] [Google Scholar]
- 49.Nomura DK, Dix MM & Cravatt BF Activity-based protein profiling for biochemical pathway discovery in cancer. Nat. Rev. Cancer 10, 630–638 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prifti E et al. A fluorogenic probe for SNAP-tagged plasma membrane proteins based on the solvatochromic molecule Nile Red. ACS Chem. Biol. 9, 606–612 (2014). [DOI] [PubMed] [Google Scholar]
Reference for Methods.
- 1.Presolski SI, Hong VP & Finn MG Copper-Catalyzed Azide-Alkyne Click Chemistry for Bioconjugation. Curr Protoc Chem Biol 3, 153–162 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhatia D, Surana S, Chakraborty S, Koushika SP & Krishnan Y A synthetic icosahedral DNA-based host-cargo complex for functional in vivo imaging. Nat Commun 2, 339 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Altschul SF, Gish W, Miller W, Myers EW & Lipman DJ Basic local alignment search tool. JMolBiol 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 4.Kamath RS & Ahringer J Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30, 313–321 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Rual J-F et al. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res 14, 2162–2168 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunbar TL, Yan Z, Balla KM, Smelkinson MG & Troemel ERC elegans detects pathogen-induced translational inhibition to activate immune signaling. Cell Host Microbe 11, 375–386 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu D, Perez RE, Rezaiekhaligh MH, Bourdi M & Truog WE Knockdown of ERp57 increases BiP/GRP78 induction and protects against hyperoxia and tunicamycin-induced apoptosis. Am J Physiol Lung Cell Mol Physiol 297, L44–51 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.