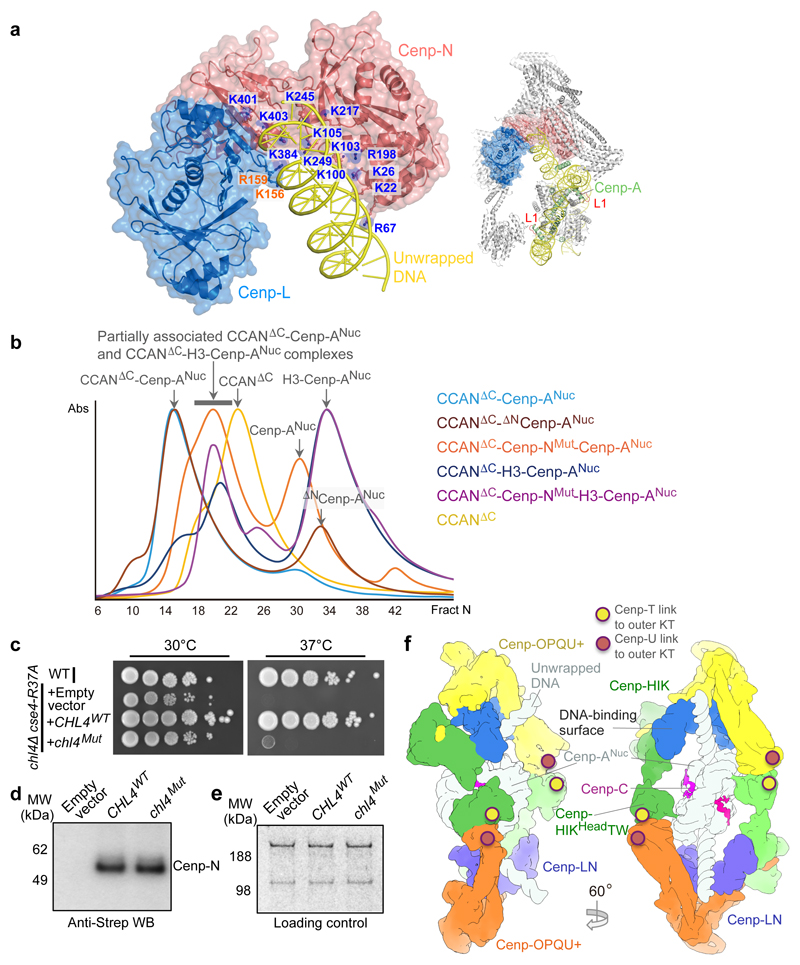
Figure 4. The Cenp-N DNA binding groove is required for stable CCAN – Cenp-ANuc interactions.
a, Surface of the Cenp-LN module showing the Cenp-N DNA binding groove engaging the unwrapped DNA, indicating the 13 mutated Arg and Lys residues of Cenp-N. Inset: overview of CCAN–Cenp-ANuc showing the Cenp-A L1 loop. b, Size exclusion chromatograms of various CCANΔCenp-C–Cenp-ANuc complexes. Wild type CCANΔCenp-C forms a complex with Cenp-ANuc, but mutating the Cenp-N DNA binding groove weakens CCAN – Cenp-ANuc interactions (Extended Data Fig. 8c, d). The binding of both CCANΔCenp-C and CCANΔCenp-C-Cenp-NMut to H3N-Cenp-ANuc is severely disrupted, with little complex formed (Extended Data Fig. 8g, h). The positions of complexes are indicated by arrows. (CCANΔC = CCANΔCenp-C). This experiment was performed independently in triplicate with similar results. c, The DNA-binding groove functions in vivo. Wild type Cenp-N (CHL4WT) rescues the growth defect of the chl4Δ cse4-R73A mutant strain at 37 °C, whereas the Cenp-NMut (chl4Mut) does not. WT: wild type strain. This experiment was performed independently ten times with similar results. d, Western blot demonstrates that Cenp-NWT and Cenp-NMut are expressed at equivalent levels in the chl4Δ cse4-R73A mutant strain. e, Loading control. Coomassie-blue stained gel shows dynein and acetyl-CoA carboxylase. Experiments in d and e were performed independently in triplicate times with similar results. f, Two views showing a representation of dimeric CCAN–Cenp-ANuc complex with the second CCAN protomer generated by the dyad symmetry of Cenp-ANuc. Sites of contact to the outer kinetochore (KT) (through Cenp-U and Cenp-T) are indicated. For gel source data see Supplementary Fig. 1.