Fig. 2.
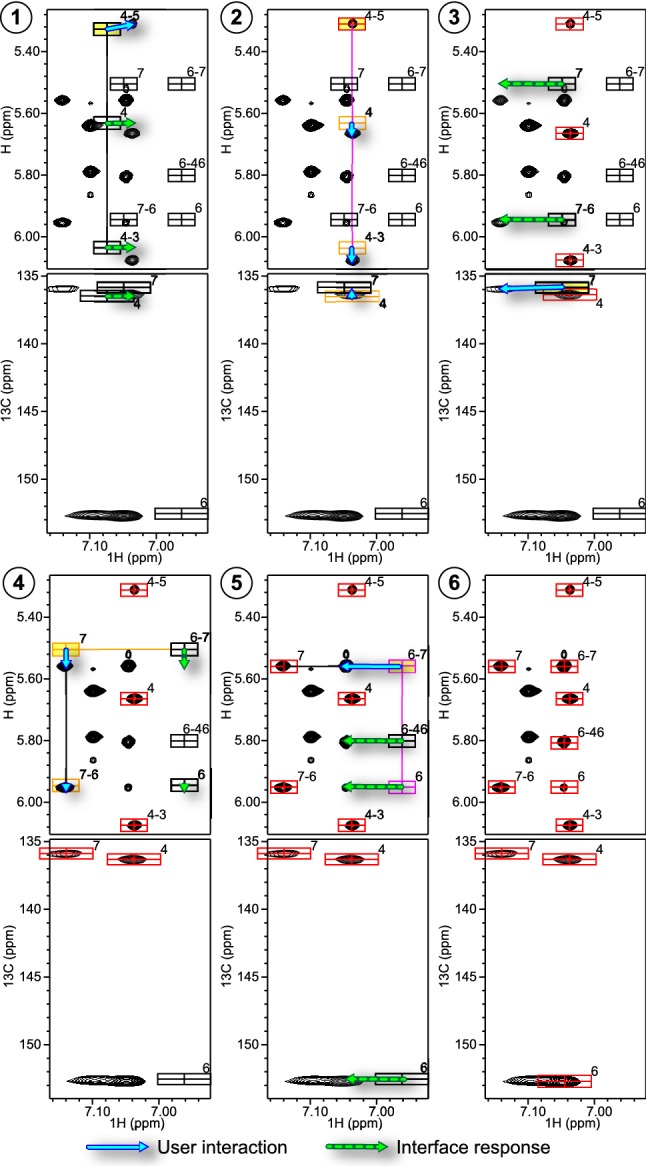
Demonstration of the assignment procedure for a portion of a 50 nt RNA. In each panel the upper spectrum is a 1H-1H NOESY and the lower a 1H-13C HMQC. 1 Peak-boxes are initially positioned according to predicted chemical shifts. Upon selecting a peak-box for positioning, the linked peak-boxes are indicated by connecting lines. Visual inspection identifies a candidate peak to which the peak-box labeled 4–5 is manually repositioned, as indicated by the solid arrow. Linked peaks are repositioned automatically, as indicated by the dashed arrows. 2 The peak-box position is frozen, indicated in red. The remaining three peak-boxes in the spin system are automatically frozen, and prevented from moving in their shared dimension, indicated in orange for the x-axis. Their associated peaks are readily identified due to this restriction. 3 Examination of the NOESY spectrum reveals two well-matched possibilities for assignment of the peak-box labeled 7. The correct assignment is found by reference to the HMQC spectrum, in which there is only one reasonable candidate. 4 Repositioning the remaining peak-boxes for the spin-system associated with this atom automatically repositions associated peak-boxes from the remaining spin-system under consideration. 5 The remaining spin-system contains peak-boxes restricted from moving along the y-axis due to previously frozen peaks, indicated in magenta, such that their associated peaks are readily identified. 6 Final positions of the peak-boxes under consideration
