Abstract
A proteome-wide protein-protein interaction (PPI) network of Methanobrevibacter ruminantium M1 (MRU), a predominant rumen methanogen, was constructed from its metabolic genes using a gene neighborhood algorithm and then compared with closely related rumen methanogens Using proteome-wide PPI approach, we constructed network encompassed 2194 edges and 637 nodes interacting with 634 genes. Network quality and robustness of functional modules were assessed with gene ontology terms. A structure-function-metabolism mapping for each protein has been carried out with efforts to extract experimental PPI concomitant information from the literature. The results of our study revealed that some topological properties of its network were robust for sharing homologous protein interactions across heterotrophic and hydrogenotrophic methanogens. MRU proteome has shown to establish many PPI sub-networks for associated metabolic subsystems required to survive in the rumen environment. MRU genome found to share interacting proteins from its PPI network involved in specific metabolic subsystems distinct to heterotrophic and hydrogenotrophic methanogens. Across these proteomes, the interacting proteins from differential PPI networks were shared in common for the biosynthesis of amino acids, nucleosides, and nucleotides and energy metabolism in which more fractions of protein pairs shared with Methanosarcina acetivorans. Our comparative study expedites our knowledge to understand a complex proteome network associated with typical metabolic subsystems of MRU and to improve its genome-scale reconstruction in the future.
Keywords: Methanobrevibacter ruminantium, Systems biology, Comparative proteomics, Network evolution, Methanogenesis, Rumen ecosystem, Methane mitigation
Highlights
-
•
A proteome-wide PPI network of Methanobrevibacter ruminantium M1 reconstructed and compared with heterotrophic and hydrogenotrophic methanogens.
-
•
Our network model gained an importance to reveal its modular network structure.
-
•
Our network model acts a computational platform for a better understanding of its cellular and metabolic behaviors.
1. Introduction
Enteric methane emission from ruminants is of great concern for its impact on global warming potential and climate changes. According to the IPCC (Intergovernmental Panel on Climate Change) report [1]; global methane emission from livestock was elevated by 11% in 2011. A loss of about 2–12% of aggregate dietary energy in ruminants can be caused by methane emission from rumen methanogens [2,3]. Several methane mitigation technologies have been developed to reduce methane emission in the gut of the ruminants. Methane mitigation interventions will make additional energy accessible to meet demand on the animal for meat and milk production and ensure the long-term sustainability of ruminant-based agriculture [4,5]. Conversely, there is no substantial strategy invented yet. Currently available anti-methanogenic compounds have been exhibiting the sign of toxic effects to beneficial rumen microbiota [[6], [7], [8], [9], [10]].
Methanobrevibacter and Methanobacterium are the predominant genera resident in the ruminants. Methanobrevibacter ruminantium M1 (MRU) is experimentally investigated to be a dominant methanogen in ruminants since it accounts for 27.3% of rumen methanogens [11]. MRU is a hydrogenotrophic rumen methanogen that uses H2 to reduce CO2 for methane biosynthesis via hydrogenotrophic methanogenesis. This organism also uses formate as a carbon source for its growth and energy metabolism [12,13]. It is the first genome sequence to be completed for rumen methanogens. This genome consists of a circular chromosome (2.93 Mbp) with 2278 coding-genes and its genome coding potential covers 144 metabolic pathways with 722 reactions, 557 enzymes, and 751 metabolites [14]. Nonetheless, its entire proteome function is still not known owing to 73% of coding-genes was annotated as hypothetical proteins and also complexity in metabolic and protein networks. Metabolic interactions between gut-microbe; microbe-microbe are imperative concerns to understand microbial mutualism in the rumen environment [10].
Genome-wide PPI networks are being prominent tools for annotation of inadequately characterized genomes in the description of the cellular machinery of prokaryotes [15]. Genome-wide PPI networks have been identified with high-throughput experimental techniques. These techniques are time-consuming and cost-effective [16,17]. Functional annotation of proteins and cognize functional network modules has been propagated by the availability of experimentally-determined PPIs and crystallographic structures of proteins in databases [[18], [19], [20]]. Most recently, a progression of computational methods has been made to predict and characterize PPI network models of several prokaryotic and eukaryotic organisms [[21], [22], [23], [24], [25]]. The functioning of molecular and metabolic pathways is highlighted with the importance of network topology, node hierarchy, and hubs, which are inclined to be crucial in protein interaction networks [26]. Hence, computational network modeling is being constructive to reveal the genotype-phenotype relationships in order to expose evolving biological properties from the genome-wide PPI networks [27,28].
The constraint of network theory is a statistical rationalization of networks and achieves similar evidence related to experimental data [29]. Understanding the interaction of proteome in genome-wide is referred to as a proteome-wide PPI network [30]. In our study, we have constructed and analyzed the network topology of a proteome-wide PPI network of MRU for a better understanding of associated metabolic subsystems. Hydrogenotrophic methanogens sustain the fractional pressures by exploiting H2 produced by cellulolytic bacteria and facilitate the bacterial fermentation in ruminants. However, metabolic interaction or cross-talk across the gut methanogens is still not studied intensively. Some network properties were predicted to reconsider their metabolic modules based on dynamic PPI networks of heterotrophic and hydrogenotrophic methanogens. Moreover, a comparative study of network stability against targeted attacks delivers a well-designed tactic. It also probed to study evolutionarily divergence of PPI network in MRU from these methanogens. Our study revealed a possible mechanism of interspecies metabolic associations of MRU with gut methanogens in the rumen ecosystem.
2. Materials and methods
2.1. Dataset
A complete genome sequence of MRU (CP001719) was retrieved from the NCBI FTP server in FASTA format (ftp://ftp.ncbi.nlm.nih.gov/). The function of this genome was annotated from the sequence by the RAST server based on the subsystems technology [31]. A metabolic network of this genome was constructed from annotated functional data by the Model SEED using a complete medium [32]. Gene set of this genome was collected from the draft metabolic network after manual validation of each gene. Gene sets of acetoclastic methanogens, Methanosarcina acetivorans C2A (MAC; AE010299) [33], M. barkeri Fusaro (MBA; CP000099) [34] and hydrogenotrophic methanogen, Methanococcus maripaludis S2 (MMP; BX950229) [35] were obtained from their genome-scale metabolic models. The proteome and metabolic information of these genomes were collected from the Kyoto Encyclopedia of Genes and Genomes (KEGG) [36] and MetaCyc [37] databases and included in the dataset of this study. Fig. 1 represents the computational process used in this study for reconstruction and comparison of the PPI network of MRU with gut methanogens.
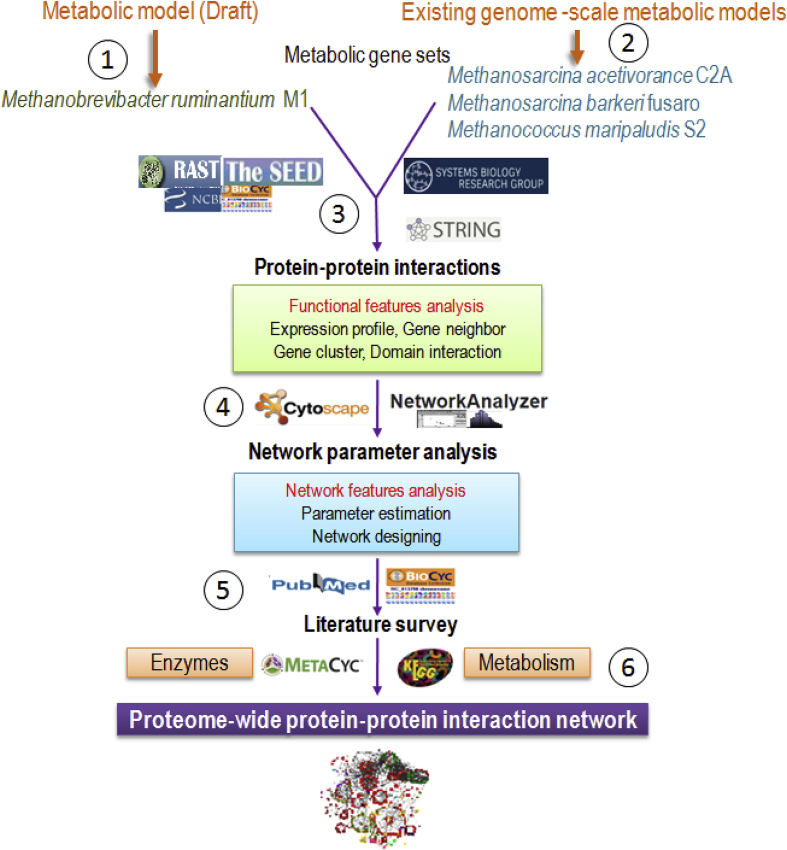
Fig. 1.
Schematic representation of the computational process used for reconstruction and comparison of the PPI network of MRU with gut methanogens. 1. Retrieval of annotated proteome information of MRU genome, 2. Retrieval of annotated proteome information of existing genome-scale models, 3. Construction of PPI network and enrichment of molecular and biological function of proteins, 4. Computational evaluation of network properties of each PPI network, 5. Network validation and functional annotation of metabolic enzymes, and 6. Comparison of differential networks of MRU with heterotrophic and hydrogenotrophic methanogens.
2.2. Network construction
To construct the PPI network of MRU, we collected experimental and predicted interacting protein pairs from the STRING 10.5 database. The current version 10.5 includes information about 9.6 million proteins from more than 4000 organisms [38]. We attempted to predict the protein interactions based on close, a co-directional neighborhood of genes in the MRU genome using a gene neighborhood algorithm [39]. This algorithm captures the functional or physical interactions between genes that are repeatedly observed in close proximity across many genomes [40]. Homomeric interactions excluded from the constructed network since it may cause bias in the subsequent analysis. We used a unique scoring framework of STRING to produce a single confidence score per prediction with a minimal false positive rate, which is particularly important to provide interaction weights. A high confidence score was set as 0.7 and more than 10 interactions included in the predicted network. It allows us to prune down the inclusive networks and ensure the accuracy of the predicted networks.
2.3. Differential network construction
We constructed the large multi-state dynamic molecular interaction networks using the DyNet module for identification and analysis of the most ‘rewired’ nodes across many network states in selected organisms [41]. We computed a dynamic rewiring score (Dn) of a node based on the variance of its corresponding row, M (p, Q, s), over the various network states, S, relative to the mean (centroid, c) as below.
The base dissimilarity measure (d) is the Euclidean distance. The vector xi represents the neighborhood M (p, Q, s) after standardization.
2.4. Network properties
The network topology is a computational measure for a quantifiable description of networks. Several network parameters have been used to describe biological phenomena characterized by distinctive network parameters and characterize the complex networks. We computed all network topological parameters of each PPI network with the Network Analysis module [42] in Cytoscape 3.4.0 [43]. Betweenness is a centrality measure of a vertex v within a graph computed in the following equation.
Where, is the number of shortest paths from s to t, and is the number of shortest paths from s to t that pass through a vertex v [44]. The path containing the least number of vertices between two vertices in a network is known as the shortest path [45]. Closeness centrality value is the inverse of the average length of the shortest paths to/from all other nodes in the graph and measures how close a node is to other nodes [46]. It was computed in the following equation.
Where, L (n, m) is the length of the shortest path between two nodes n and m. The average clustering coefficient for all nodes in a network was taken to characterize the overall tendency of nodes to form clusters or groups as below.
Where, kn is the number of neighbors of n and en is the number of connected pairs between all neighbors of n [47]. The topological coefficient is a relative measure for the extent to which a node shares neighbors kn with other nodes n defined in the following equation.
Where, J(n,m) is defined for all nodes m that share at least one neighbor with n [48]. The characteristic path length measures the typical separation between two vertices in the network [45]. The network diameter d is the shortest path between any two nodes in a network, which defined in the following equation.
Where, dG (u, v) is the shortest path between u and v in G. The number of interacting partner proteins and its distribution allows distinctive between various network topologies is defined as the node degree [49]. We also computed other network parameters such as in-degree, out-degree, neighborhood connectivity, and stress centrality as methods described earlier [44,50].
2.5. Network validation
We validated each predicted PPI network by random selection using the iLOOP server [51]. Gene sets in each network were compared with the semantic similarities of gene ontology (GO) terms between constructed PPI and random networks and then confirmed the functional similarities of proteins [24]. We captured enriched molecular and metabolic functions of proteins in each network from functional classification systems in GO and Pfam [52]. The homologous function of proteins in each PPI network was searched out and then enriched to GO terms for the detection of associated metabolic subsystems using different database searching. NCBI-PubMed (www.ncbi.nlm.nih.gov/pubmed/), MetaCyc [37], KEGG (Kanehisa and Goto, 2018), ModelSEED [53], PATRIC [54] and BRENDA [55] were the databases used for this purpose.
3. Results
3.1. Construction of PPI network of MRU
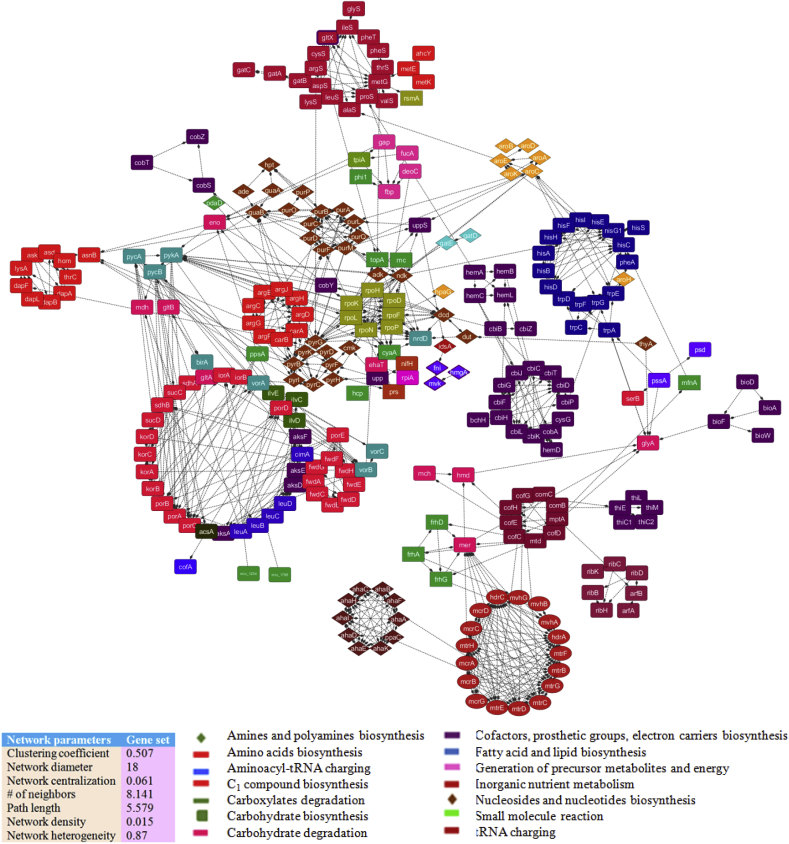
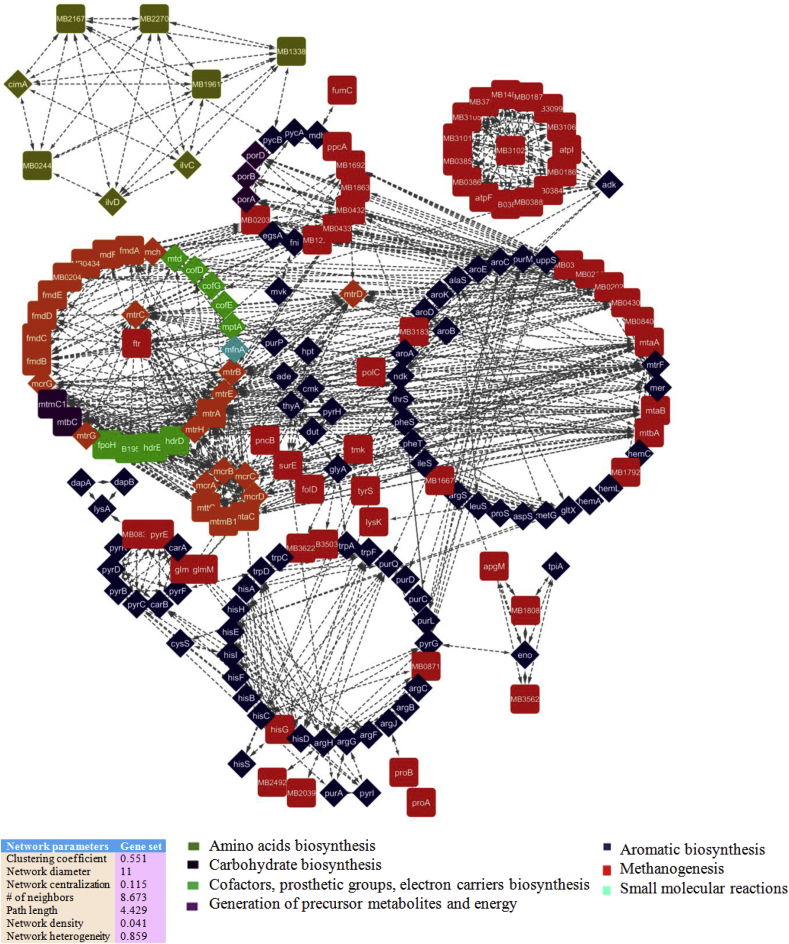
As shown in Fig. 2, a proteome-wide PPI network of MRU was reconstructed from a metabolic gene set containing 634 genes. The proteins are directly interconnected in the metabolic network of a genome. Therefore, metabolic genes are only included in the datasets of this study. The core network consists of 637 nodes and 2194 edges with a high betweenness centrality value. It was evaluated with network parameters including a quantify modularity, clustering coefficient (0.507), diameter (18), centralization (0.061), path length (5.579), density (0.015), and network heterogeneity (0.87). Each of the network properties was appreciably enriched to describe metabolic functions over-represented by clustered nodes. It was further validated with well-characterized crystallographic structures available in the protein structural databases but structural information to the target proteins is limited to date. Apart from the available crystallographic structures, the proteins in the network were compared with gene ontology terms for functional assignment. The reliability of the predicted PPI network was evaluated from the structure-function-metabolism links of interacting proteins. Network topology analysis indicates the constructed network has more influence over the information flow in the network as a node with a higher betweenness centrality value (Table 1; Table S1). The distribution of closeness and betweenness centrality value is ranged from 0.1 to 0.2, representing the number of shortest paths passing through a specific node in the network. The overall network parameters indicate that the PPI network model of this genome is more stable, consistent and conserved across the modules.
Fig. 2.
A core proteome-wide protein interaction network of MRU. Its whole proteome network was constructed from the gene set (634 genes) of a draft metabolic model of MRU using a neighborhood algorithm. The draft metabolic network was constructed from the Model SEED. The core network consisted of 637 nodes and 2194 edges with a high betweenness centrality value and 2194 edges.
Table 1.
Comparison of differential proteome-wide protein interaction network parameters between MRU and existing genome-scale metabolic models of methanogenic archaea. Network radius and network density were calculated as 1 and 0, respectively.
| Network parameters | Dynamic protein networks |
Differential protein networks |
||||||
|---|---|---|---|---|---|---|---|---|
| MRU | MAC | MBA | MMP | MRU | MAC | MBA | MMP | |
| Clustering coefficient | 0.288 | 0.279 | 0.276 | 0.288 | 0.182 | 0.242 | 0.235 | 0.252 |
| Connected components | 15 | 16 | 12 | 15 | 11 | 21 | 16 | 12 |
| Network diameter | 22 | 23 | 22 | 22 | 8 | 9 | 24 | 27 |
| Path length | 7.173 | 7.189 | 7.813 | 7.173 | 2.17 | 7.429 | 8.532 | 7.745 |
| # of neighbors | 7.693 | 10.471 | 7.911 | 7.693 | 6.319 | 10.061 | 7.211 | 7.085 |
| # of nodes | 462 | 594 | 384 | 462 | 426 | 590 | 380 | 457 |
The functional similarities of interacting proteins with the same topology calculated according to their GO semantic similarities. GO enrichment analysis for the important nodes was performed to investigate network-associated metabolism. It described many protein pairs within the same functional categories in the PPI network of MRU. We have categorized, grouped and aligned all interacting proteins and associated metabolism (Fig. 2). It indicates more fractions of coding-genes found to be associated with biosynthesis of cofactors, prosthetic groups, electron carriers (125) and generation of precursor metabolites and energy (103). Above 80 coding-genes are accounted for the biosynthesis of amino acids, nucleosides, and nucleotides. Aminoacyl-tRNA charging, carboxylates degradation, carbohydrate metabolism and biosynthesis of fatty acid, and lipids are associated with 30 coding-genes in the PPI network of this organism.
3.2. Differential PPI network between MRU and MAC
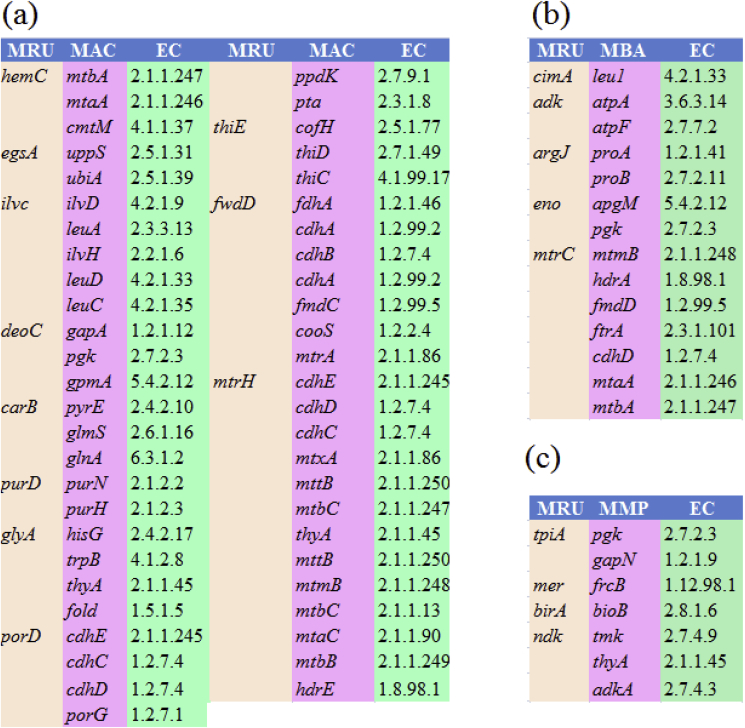
We constructed a differential proteome-wide PPI network from the gene sets of MRU and MAC to find out homologs protein pairs between them. This network model consists of 466 nodes and 2513 edges with 410 interacting proteins (Fig. 3). Estimates of network properties show that it has a 0.537 clustering coefficient, 14 diameters, 0.026 density, 0.106 centralizations, and 0.987 heterogeneity. A large fraction of coding-genes from their protein pairs are associated with amino acid metabolism (206), C1 compounds assimilation (129), precursor metabolites and energy (70). The significant fraction of their interacting proteins accounts for the biosynthesis of cofactors, prosthetic groups, electron carriers (48), small molecule reactions (57), and cell structures metabolism (37). It implies that both genomes regulate the interacting protein modules with similar transcription and metabolic patterns through conserved PPI networks. The highly connected hub proteins in these metabolic subsystems are evolutionarily conserved between MRU and MAC. Besides, we predicted new protein interactions and proteins with unknown function from its conserved network modules. Shown by our comparative analysis, we predicted 11 coding-genes (hemC, egsA, ilvc, deoC, carB, purD, glyA, porD, thiE, fwdD and mtrH) from MRU exhibiting to interact with 54 coding-genes from MAC (Fig. 4a). These coding-genes form the sub-protein networks separately from a differential PPI network of MRU and MAC. It suggests that those genes are originated from a common ancestor and functionally shared together to make a conserved network module in each genome for amino acid and energy metabolism. It provides a clue to the understanding of complex regulatory and metabolic networks of MRU.
Fig. 3.
DyNet visualization of the dynamic proteome-wide PPI network constructed by combining the gene sets (410 genes) collected from the genome-scale metabolic models of MRU and MAC using a neighborhood algorithm. The core network consisted of 466 nodes and 2513 edges with a high betweenness centrality value. Diamond and circle shapes represent the proteins from MRU and MAC, respectively.
Fig. 4.
Functional prediction of gene homologs identified from MRU using interacting proteins in differential PPI networks of MRU and MAC (a); MRU and MBA (b); MRU and MMP (c).
3.3. Differential PPI network between MRU and MBA
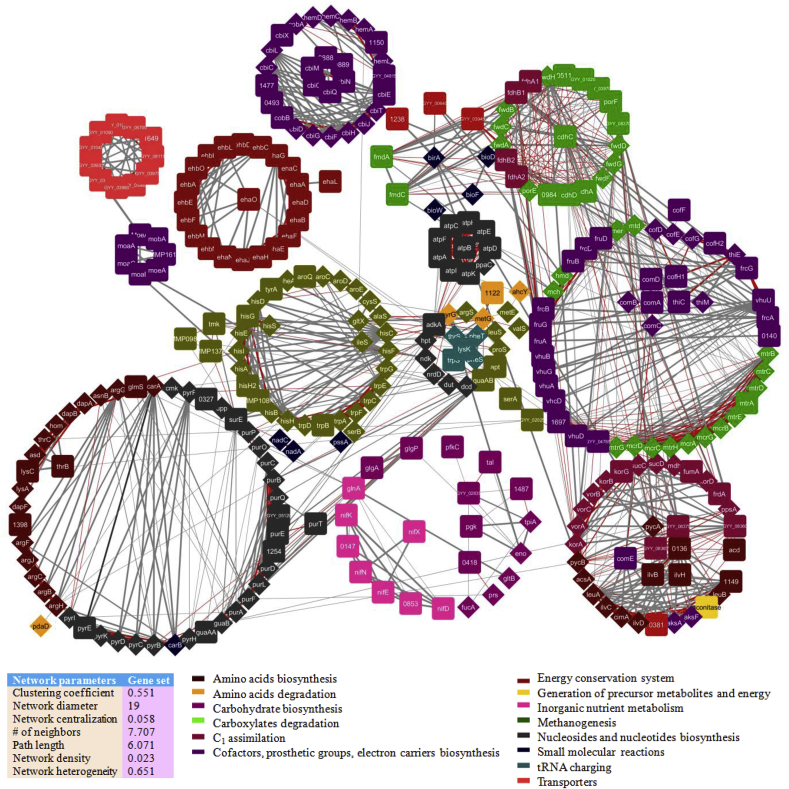
As similar to MAC, a differential proteome-wide PPI network constructed for MRU and MBA (Fig. 5). It contains 466 nodes and 2470 edges with 432 interacting proteins. We computed the clustering coefficient (0.551), diameter (11), density (0.041), centralization (0.115), and heterogeneity (0.859), which reflects the stability and consistency of predicted PPI network. The biosynthesis of amino acids (115) and carbohydrates (76) and methanogenesis (58) are major metabolic pathways associating with protein pairs predicted from this network. Many metabolic subsystems have shown a low fraction of coding-genes to be interacted in core PPI networks and or to make conserved hubs of these genomes. As shown in Fig. 4b, cimA, adk, argJ, eno, and mtrC genes from MRU are differentially interacted with 14 coding genes of MBA and formed 5 sub-networks in this network. All of these interacting proteins are shared between MRU and MBA and found to mediate mainly hydrogenotrophic and carboxydotrophic methanogenesis. Perhaps, it suggests the ability of this organism to grow on carbon monoxide as a carbon source apart from CO2 reduction and formate oxidation under selective pressure.
Fig. 5.
DyNet visualization of the dynamic proteome-wide PPI network constructed by combining the gene sets (432 genes) collected from the genome-scale metabolic models of MRU and MBA using a neighborhood algorithm. The core network consisted of 466 nodes and 2470 edges with a high betweenness centrality value. Diamond and circle shapes represent the proteins from MRU and MBA, respectively.
3.4. Differential PPI network between MRU and MMP
For comparing MRU with MMP, we constructed a differential proteome-wide PPI network consisting of 371 nodes and 1278 edges with 348 interacting proteins (Fig. 6). Network parameters including clustering coefficient (0.551), network diameter (19), centralization (0.058), density (0.023) and heterogeneity (0.651) were calculated for this network. It represents the robustness and consistency of network with more biological and statistical significance. Analysis of interacting proteins shows that a large fraction of protein pairs are accounted for the biosynthesis of amino acids (164), nucleosides and nucleotides (51), and C1 compounds assimilation (77). Some of the coding genes (tpiA, mer/mtd, birA, and ndk) from MRU are homologs to those genes found in MMP (Fig. 4c). We found 8 gene homologs from MMP showing to interact with 4 coding-genes of MRU. All of these coding genes are contributed to the biosynthesis of nucleosides and nucleotides in both genomes. As a result of our study, we suggest that despite hydrogenotrophic methanogenesis, both genomes are shared some gene clusters (pyr, car, pur, gua, cmk, atp, adk, and ndk) for nucleic acid metabolism.
Fig. 6.
DyNet visualization of the dynamic proteome-wide PPI network constructed by combining the gene sets (348 genes) collected from the genome-scale metabolic models of MRU and MMP using a neighborhood algorithm. The core network consisted of 371 nodes and 1287 edges with a high betweenness centrality value. Diamond and circle shapes represent the proteins from MRU and MMP, respectively.
4. Discussion
Yeast two-hybrid system and tandem affinity purification are time-consuming and cost-effective high-throughput experimental methods for the construction of a large number of PPI networks [16,17]. Functional annotation and assignments of proteins has been improved advance by available experimental data on PPIs and crystallographic structures in the public domain databases [18]. Experimental and computational approaches have been utilized to drive the molecular hypothesis of several organisms from their proteome-wide PPI networks [23,24]. Therefore, we developed a proteome-wide PPI network model for MRU consisting of 2194 edges, 637 nodes, and 634 interacting genes. Estimates of its network parameters and topology have characterized this network model, which supported us to reveal our molecular hypothesis of this organism at the proteome-scale. The PPI networks constructed from this study were experimentally validated with relevant structures, COG and GO terms [17,29,30,56]. The results of our study indicated its species-specific network characteristics, which are associated with distinctive metabolic subsystems. It also pointed out that MRU proteome has shared homologous features mainly with metabolic enzymes of MAC compared to MBA and MMP proteomes. Some poorly characterized proteins and their complexes were functionally annotated from interacting proteins with a known function from differential PPI networks of other organisms.
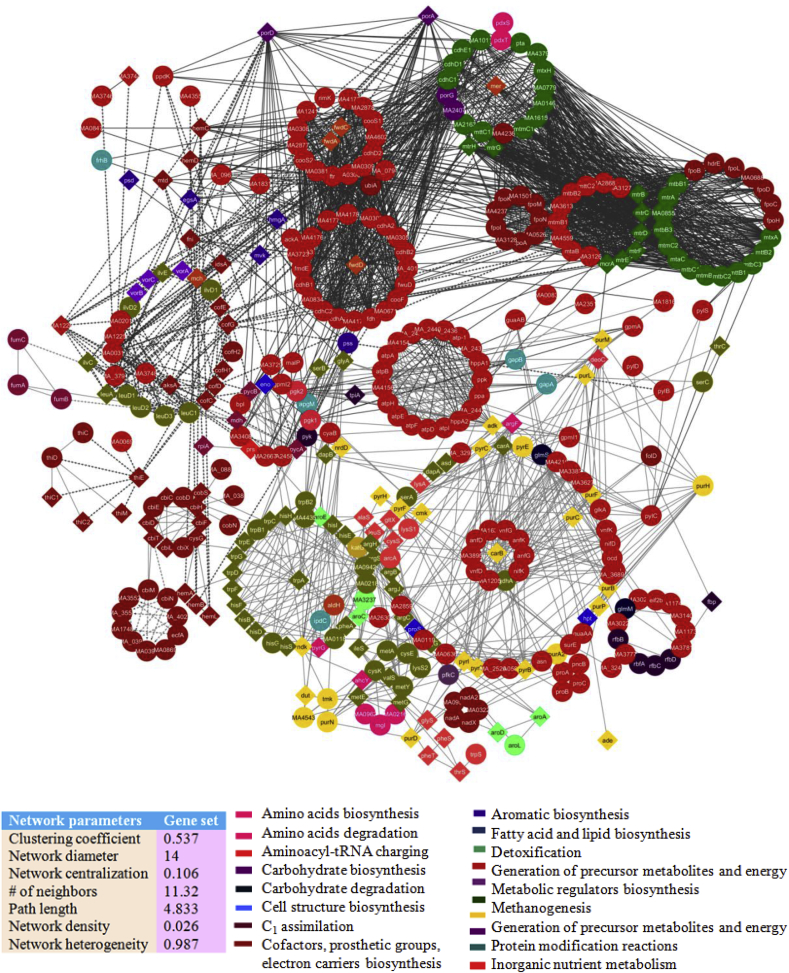
Computational prediction and characterization of PPI networks have been carried out for some bacteria including Escherichia coli [57], Helicobacter pylori [58], Mycobacterium tuberculosis [59], Bacillus subtilis [60], Bacillus licheniformis [24], Synechocystis sp. PCC 6803 [21]. Several investigators have been reconstructed PPI networks of few eukaryotic organisms including Tetraselmis subcordiformis [22], Candida albicans, Aspergillums fumigatus [23] and methanogenic archaean, Methanothermobacter thermautotrophicus [19]. Hence, in silico methods are more reliable, accurate and consistent for these PPI network models as similar evidence related to experimental data. Nevertheless, no one is reported a large proteome-wide PPIs of archaea yet for studying the basic genome biology and lifestyle in the rumen ecosystem. A proteome-wide PPI network constructed from this study was the first computational model of the archaeal domain. It described significant interacting protein pairs associated with central metabolic networks of MRU. New protein pairs or function of uncharacterized proteins have been disclosed from conserved network modules and highly connected hubs. As shown by our analysis, the present network models could serve as a computational platform to describe its growth physiology in the gut of ruminants. Another one noteworthy is that PPI network models constructed from the metabolic gene sets of the rumen, heterotrophic and hydrogenotrophic methanogens. As a result, gene sets derived from the genome-scale metabolic networks allow us to infer underlying cellular metabolism associated with PPI sub-networks of rumen methanogens.
Studying network topology and node hierarchy, the centrality parameter is being a major concern to determine the importance of each node in a network [26]. It described evolving properties and the functioning molecular pathways of the PPI network in MRU. The term degree distribution represents the number of proteins that interact with another specific protein. The degree distribution was slowly decreased in interacting proteins, leading to the generation of a network pattern, as calculated to the earlier models [27,28]. As the results of network parameters, the PPI network model of MRU has attained a general characteristic to best fit in all currently available PPI networks and its topological coefficient also perfectly followed a power law distribution. GO enrichment analysis indicated that the function of interacting proteins from MRU directly linked to central metabolism of respective PPI networks of MMA. Topological robustness analysis probed the divergence and conservation of modularity of predicted PPI networks between heterotrophic and hydrogenotrophic methanogens, as described earlier [41].
Results of metabolic analysis show, that biosynthesis of cofactors, prosthetic groups, electron carriers, generation of precursor metabolites and energy associated with more fractions of interacting proteins in MRU. The proteins pairs predicted from a differential network of MRU and MAC have shown more fractions of them contributed to amino acid and energy metabolism. As shown by our analysis, MRU has the ability to grow on carbon monoxide via carboxydotrophic methanogenesis at the specific rumen environmental niche [2]. Both MRU and MMP found to share some interacting proteins commonly for the biosynthesis of nucleosides and nucleotides. As a result of this study, it is a hypothesis that interacting proteins from each genome are making a sub-protein network from core PPI network to a particular metabolic subsystem so as to withstand their growth in the rumen environment. Such interacting proteins and networks might have evolved from a common ancestor and they shared together to make a conserved functional network module in each genome.
5. Conclusions
We reconstructed a proteome-wide PPI network for MRU comprising 2194 edges and 637 nodes interacting with 634 genes of MRU. GO terms of functional modules assessed with network quality and robustness. The great efforts have been made to extract experimental PPI concomitant information from the literature for the structure-function-metabolism mapping. Many network characteristics are shared with heterotrophic and hydrogenotrophic methanogens.
Genes involved in hydrogenotrophic methanogenesis are shared commonly across the MRU, MAC, MBA and MMP but genes involved in acetoclastic methanogenesis are absent in MMP. MRU has the ability to grow and survive in the rumen environment by the establishment of metabolic subsystems including biosynthesis of amino acids, nucleosides, nucleotides, and energy metabolism through PPI sub-networks. The network properties of this model have shown a good agreement with experimental data and the previous protein networks. Moreover, we suggest the network simulations check the network stability, implicating a potential molecular basis for the stochastic nature of methanogenesis. It is also important to note that protein structural and gene expression data will improve the accuracy and biological reliability of this model to present the molecular hypothesis on methane mitigation mechanism of rumen methanogens.
Declaration of competing interest
The authors declare that they have no competing interests.
Acknowledgments
The authors would like to thank the University Grants Commission (RA-2012-14-SC-TAM-1768), New Delhi, India for financial assistance.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2019.100698.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.IPCC (Intergovernmental Panel on Climate change) 2007. Climate Change 2007: the Physical Science Basis, Contribution of Working Group I to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change. [Google Scholar]
- 2.Hook S.E., Wright A.D., McBride B.W. Methanogens: methane producers of the rumen and mitigation strategies. Archaea. 2010;2010:945785. doi: 10.1155/2010/945785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hristov A.N., Oh J., Firkins J.L., Dijkstra J., Kebreab E., Waghorn G., Makkar H.P., Adesogan A.T., Yang W., Lee C., Gerber P.J., Henderson B., Tricarico J.M. Special topics-Mitigation of methane and nitrous oxide emissions from animal operations: I. A review of enteric methane mitigation options. J. Anim. Sci. 2013;91:5045–5069. doi: 10.2527/jas.2013-6583. [DOI] [PubMed] [Google Scholar]
- 4.Thornton P.K. Livestock production: recent trends, future prospects. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2010;365:2853–2867. doi: 10.1098/rstb.2010.0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thornton P.K., Herrero M. Potential for reduced methane and carbon dioxide emissions from livestock and pasture management in the tropics. Proc. Natl. Acad. Sci. U.S.A. 2010;107:19667–19672. doi: 10.1073/pnas.0912890107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reynolds C.K., Humphries D.J., Kirton P., Kindermann M., Duval S., Steinberg W. Effects of 3-nitrooxypropanol on methane emission, digestion, and energy and nitrogen balance of lactating dairy cows. J. Dairy Sci. 2014;97:3777–3789. doi: 10.3168/jds.2013-7397. [DOI] [PubMed] [Google Scholar]
- 7.Hristov A.N., Oh J., Giallongo F., Frederick T.W., Harper M.T., Weeks H.L., Branco A.F., Moate P.J., Deighton M.H., Williams S.R., Kindermann M., Duval S. An inhibitor persistently decreased enteric methane emission from dairy cows with no negative effect on milk production. Proc. Natl. Acad. Sci. U.S.A. 2015;112:10663–10668. doi: 10.1073/pnas.1504124112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lopes J.C., de Matos L.F., Harper M.T., Giallongo F., Oh J., Gruen D., Ono S., Kindermann M., Duval S., Hristov A.N. Effect of 3-nitrooxypropanol on methane and hydrogen emissions, methane isotopic signature, and ruminal fermentation in dairy cows. J. Dairy Sci. 2016;99:5335–5344. doi: 10.3168/jds.2015-10832. [DOI] [PubMed] [Google Scholar]
- 9.Chellapandi P., Bharathi M., Prathiviraj R., Sasikala R., Vikraman M. Genome-scale metabolic model as a virtual platform to reveal the environmental contribution of methanogens. Curr. Biotechnol. 2017;6 [Google Scholar]
- 10.Chellapandi P., Bharathi M., Sangavai C., Prathiviraj R. Methanobacterium formicicum as a target rumen methanogen for the development of new methane mitigation interventions: A review. Vet Anim Sci. 2018;6:86–94. doi: 10.1016/j.vas.2018.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Janssen P.H., Kirs M. Structure of the archaeal community of the rumen. Appl. Environ. Microbiol. 2008;74:3619–3625. doi: 10.1128/AEM.02812-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zabranska J., Pokorna D. Bioconversion of carbon dioxide to methane using hydrogen and hydrogenotrophic methanogens. Biotechnol. Adv. 2018;36:707–720. doi: 10.1016/j.biotechadv.2017.12.003. [DOI] [PubMed] [Google Scholar]
- 13.Bharathi M., Chellapandi P. Intergenomic evolution and metabolic cross-talk between rumen and thermophilic autotrophic methanogenic archaea. Mol. Phylogenetics Evol. 2017;107:293–304. doi: 10.1016/j.ympev.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 14.Leahy S.C., Kelly W.J., Altermann E., Ronimus R.S., Yeoman C.J., Pacheco D.M., Li D., Kong Z., McTavish S., Sang C., Lambie S.C., Janssen P.H., Dey D., Attwood G.T. The genome sequence of the rumen methanogen Methanobrevibacter ruminantium reveals new possibilities for controlling ruminant methane emissions. PLoS One. 2010;5:e8926. doi: 10.1371/journal.pone.0008926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mostafavi S., Ray D., Warde-Farley D., Grouios C., Morris Q. GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 2008;9:S4. doi: 10.1186/gb-2008-9-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Browne F., Wang H., Zheng H., Azuaje F. A knowledge-driven probabilistic framework for the prediction of protein-protein interaction networks. Comput. Biol. Med. 2010;40:306–317. doi: 10.1016/j.compbiomed.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 17.Raman K. Construction and analysis of protein-protein interaction networks. Autom. Exp. 2010;2:2. doi: 10.1186/1759-4499-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zahiri J., Bozorgmehr J.H., Masoudi-Nejad A. Computational prediction of protein-protein interaction networks: Algorithms and resources. Curr. Genom. 2013;14:397–414. doi: 10.2174/1389202911314060004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prathiviraj R., Chellapandi P. Functional annotation of operome from Methanothermobacter thermautotrophicus ΔH: An insight to metabolic gap filling. Int. J. Biol. Macromol. 2019;123:350–362. doi: 10.1016/j.ijbiomac.2018.11.100. [DOI] [PubMed] [Google Scholar]
- 20.Sangavai C., Prathiviraj R., Chellapandi P. Functional prediction, characterization, and categorization of operome from Acetoanaerobium sticklandii DSM 519. Anaerobe. 2019;2019:102088. doi: 10.1016/j.anaerobe.2019.102088. [DOI] [PubMed] [Google Scholar]
- 21.Li Y., Rao N., Yang F., Zhang Y., Yang Y., Liu H.M., Guo F., Huang J. Biocomputational construction of a gene network under acid stress in Synechocystis sp. PCC 6803. Res. Microbiol. 2014;165:420–428. doi: 10.1016/j.resmic.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 22.Ji C., Cao X., Yao C., Xue S., Xiu Z. Protein-protein interaction network of the marine microalga Tetraselmis subcordiformis: prediction and application for starch metabolism analysis. J. Ind. Microbiol. Biotechnol. 2014;41:1287–1296. doi: 10.1007/s10295-014-1462-z. [DOI] [PubMed] [Google Scholar]
- 23.Remmele C.W., Luther C.H., Balkenhol J., Dandekar T., Müller T., Dittrich M.T. Integrated inference and evaluation of host-fungi interaction networks. Front. Microbiol. 2015;6:764. doi: 10.3389/fmicb.2015.00764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han Y.C., Song J.M., Wang L., Shu C.C., Guo J., Chen L.L. Prediction and characterization of protein-protein interaction network in Bacillus licheniformis WX-02. Sci. Rep. 2016;6:19486. doi: 10.1038/srep19486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prathiviraj R., Berchmans Sheela, Chellapandi P. Analysis of modularity in proteome-wide protein interaction networks of Methanothermobacter thermautotrophicus strain ΔH and metal-loving bacteria. J. Protein Proteom. 2019 [Google Scholar]
- 26.Azevedo H., Moreira-Filho C.A. Topological robustness analysis of protein interaction networks reveals key targets for overcoming chemotherapy resistance in glioma. Sci. Rep. 2015;5:16830. doi: 10.1038/srep16830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carreno-Quintero N., Bouwmeester H.J., Keurentjes J.J. Genetic analysis of metabolome-phenotype interactions: from model to crop species. Trends Genet. 2013;29:41–50. doi: 10.1016/j.tig.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 28.Liu Y.Y., Slotine J.J., Barabási A.L. Control centrality and hierarchical structure in complex networks. PLoS One. 2012;7:e44459. doi: 10.1371/journal.pone.0044459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Christakis N.A., Fowler J.H. Social contagion theory: examining dynamic social networks and human behavior. Stat. Med. 2013;32:556–577. doi: 10.1002/sim.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hao T., Peng W., Wang Q., Wang B., Sun J. Reconstruction and application of protein-protein interaction network. Int. J. Mol. Sci. 2016;17:E907. doi: 10.3390/ijms17060907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aziz R.K., Bartels D., Best A.A., DeJongh M., Disz T., Edwards R.A., Formsma K., Gerdes S., Glass E.M., Kubal M., Meyer F., Olsen G.J., Olson R., Osterman A.L., Overbeek R.A., McNeil L.K., Paarmann D., Paczian T., Parrello B., Pusch G.D., Reich C., Stevens R., Vassieva O., Vonstein V., Wilke A., Zagnitko O. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Devoid S., Overbeek R., DeJongh M., Vonstein V., Best A.A., Henry C. Automated genome annotation and metabolic model reconstruction in the SEED and Model SEED. Methods Mol. Biol. 2013;985:17–45. doi: 10.1007/978-1-62703-299-5_2. [DOI] [PubMed] [Google Scholar]
- 33.Benedict M.N., Gonnerman M.C., Metcalf W.W., Price N.D. Genome-scale metabolic reconstruction and hypothesis testing in the methanogenic archaeon Methanosarcina acetivorans C2A. J. Bacteriol. 2012;194:855–865. doi: 10.1128/JB.06040-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gonnerman M.C., Benedict M.N., Feist A.M., Metcalf W.W., Price N.D. Genomically and biochemically accurate metabolic reconstruction of Methanosarcina barkeri Fusaro, iMG746. Biotechnol. J. 2013;8:1070–1079. doi: 10.1002/biot.201200266. [DOI] [PubMed] [Google Scholar]
- 35.Goyal N., Widiastuti H., Karimi I.A., Zhou Z. A genome-scale metabolic model of Methanococcus maripaludis S2 for CO2 capture and conversion to methane. Mol. Biosyst. 2014;10:1043–1054. doi: 10.1039/c3mb70421a. [DOI] [PubMed] [Google Scholar]
- 36.Kanehisa M., Sato Y., Furumichi M., Morishima K., Tanabe M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2018 doi: 10.1093/nar/gky962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caspi R., Billington R., Fulcher C.A., Keseler I.M., Kothari A., Krummenacker M., Latendresse M., Midford P.E., Ong Q., Ong W.K., Paley S., Subhraveti P., Karp P.D. The MetaCyc database of metabolic pathways and enzymes. Nucleic Acids Res. 2018;46:D633–D639. doi: 10.1093/nar/gkx935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P., Jensen L.J., von Mering C. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Snel B., Lehmann G., Bork P., Huynen M.A. STRING: a web-server to retrieve and display the repeatedly occurring neighborhood of a gene. Nucleic Acids Res. 2000;28:3442–3444. doi: 10.1093/nar/28.18.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yanai I., DeLisi C. The society of genes: networks of functional links between genes from comparative genomics. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-11-research0064. research0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goenawan I.H., Bryan K., Lynn D.J. DyNet: visualization and analysis of dynamic molecular interaction networks. Bioinformatics. 2016;32:2713–2715. doi: 10.1093/bioinformatics/btw187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Assenov Y., Ramírez F., Schelhorn S.E., Lengauer T., Albrecht M. Computing topological parameters of biological networks. Bioinformatics. 2008;24:282–284. doi: 10.1093/bioinformatics/btm554. [DOI] [PubMed] [Google Scholar]
- 43.Doncheva N.T., Assenov Y., Domingues F.S., Albrecht M. Topological analysis and interactive visualization of biological networks and protein structures. Nat. Protoc. 2012;7:670–685. doi: 10.1038/nprot.2012.004. [DOI] [PubMed] [Google Scholar]
- 44.Brandes U. A faster algorithm for betweenness centrality. J. Math. Sociol. 2001;25:163–177. [Google Scholar]
- 45.Watts D.J., Strogatz S.H. Collective dynamics of 'small-world' networks. Nature. 1998;393:440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- 46.Newman M.E.J. A measure of betweenness centrality based on random walks. 2003. 0309045
- 47.Barabási A.L., Oltvai Z.N. Network biology: understanding the cell's functional organization. Nat. Rev. Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 48.Stelzl U., Worm U., Lalowski M., Haenig C., Brembeck F.H., Goehler H., Stroedicke M., Zenkner M., Schoenherr A., Koeppen S., Timm J., Mintzlaff S., Abraham C., Bock N., Kietzmann S., Goedde A., Toksöz E., Droege A., Krobitsch S., Korn B., Birchmeier W., Lehrach H., Wanker E.E. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 49.Ravasz E., Somera A.L., Mongru D.A., Oltvai Z.N., Barabási A.L. Hierarchical organization of modularity in metabolic networks. Science. 2002;297:1551–1555. doi: 10.1126/science.1073374. [DOI] [PubMed] [Google Scholar]
- 50.Maslov S., Sneppen K. Specificity and stability in topology of protein networks. Science. 2002;296:910–913. doi: 10.1126/science.1065103. [DOI] [PubMed] [Google Scholar]
- 51.Planas-Iglesias J., Marin-Lopez M.A., Bonet J., Garcia-Garcia J., Oliva B. iLoops: a protein-protein interaction prediction server based on structural features. Bioinformatics. 2013;29:2360–2362. doi: 10.1093/bioinformatics/btt401. [DOI] [PubMed] [Google Scholar]
- 52.Finn R.D., Bateman A., Clements J., Coggill P., Eberhardt R.Y., Eddy S.R., Heger A., Hetherington K., Holm L., Mistry J., Sonnhammer E.L.L., Tate J., Punta M. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Henry C.S., DeJongh M., Best A.A., Frybarger P.M., Linsay B., Stevens R.L. High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat. Biotechnol. 2010;28:977–982. doi: 10.1038/nbt.1672. [DOI] [PubMed] [Google Scholar]
- 54.Gillespie J.J., Wattam A.R., Cammer S.A., Gabbard J.L., Shukla M.P., Dalay O., Driscoll T., Hix D., Mane S.P., Mao C., Nordberg E.K., Scott M., Schulman J.R., Snyder E.E., Sullivan D.E., Wang C., Warren A., Williams K.P., Xue T., Yoo H.S., Zhang C., Zhang Y., Will R., Kenyon R.W., Sobral B.W. PATRIC: the comprehensive bacterial bioinformatics resource with a focus on human pathogenic species. Infect. Immun. 2011;79:4286–4298. doi: 10.1128/IAI.00207-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schomburg I., Chang A., Ebeling C., Gremse M., Heldt C., Huhn G., Schomburg D. BRENDA, the enzyme database: updates and major new developments. Nucleic Acids Res. 2004;32:D431–D433. doi: 10.1093/nar/gkh081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taylor I.W., Linding R., Warde-Farley D., Liu Y., Pesquita C., Faria D., Bull S., Pawson T., Morris Q., Wrana J.L. Dynamic modularity in protein interaction networks predicts breast cancer outcome. Nat. Biotechnol. 2009;27:199–204. doi: 10.1038/nbt.1522. [DOI] [PubMed] [Google Scholar]
- 57.Rajagopala S.V., Sikorski P., Kumar A., Mosca R., Vlasblom J., Arnold R., Franca-Koh J., Pakala S.B., Phanse S., Ceol A., Häuser R., Siszler G., Wuchty S., Emili A., Babu M., Aloy P., Pieper R., Uetz P. The binary protein-protein interaction landscape of Escherichia coli. Nat. Biotechnol. 2014;32:285–290. doi: 10.1038/nbt.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Häuser R., Ceol A., Rajagopala S.V., Mosca R., Siszler G., Wermke N., Sikorski P., Schwarz F., Schick M., Wuchty S., Aloy P., Uetz P. A second-generation protein-protein interaction network of Helicobacter pylori. Mol. Cell. Proteom. 2014;13:1318–1329. doi: 10.1074/mcp.O113.033571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huo T., Liu W., Guo Y., Yang C., Lin J., Rao Z. Prediction of host-pathogen protein interactions between Mycobacterium tuberculosis and Homo sapiens using sequence motifs. BMC Bioinf. 2015;16:100. doi: 10.1186/s12859-015-0535-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marchadier E., Carballido-López R., Brinster S., Fabret C., Mervelet P., Bessières P., Noirot-Gros M.F., Fromion V., Noirot P. An expanded protein-protein interaction network in Bacillus subtilis reveals a group of hubs: Exploration by an integrative approach. Proteomics. 2011;11:2981–2991. doi: 10.1002/pmic.201000791. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.