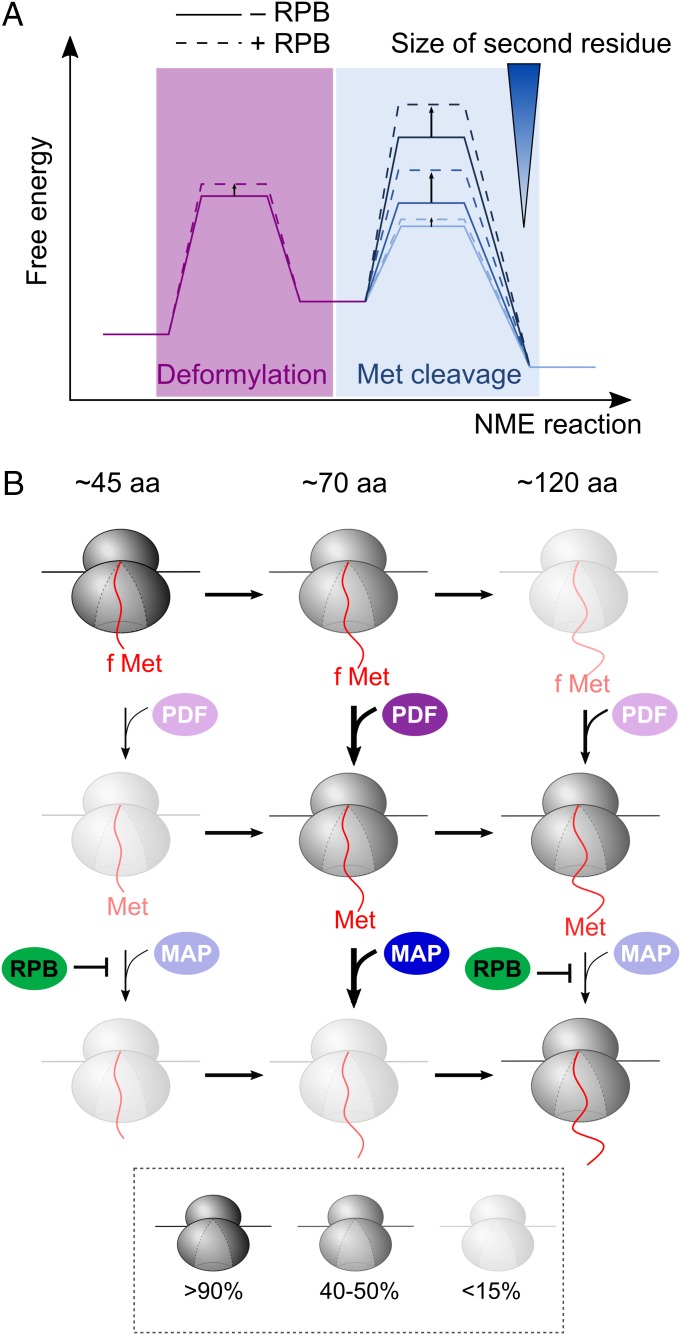
Fig. 6.
Model for cotranslational NME mediated by PDF and MAP. (A) Free energy profiles to depict the change in rate-limiting step during cotranslational NME due to both the sequence specificity of MAP and regulation by RPBs. As the second residue on the nascent protein becomes larger, the rate-limiting step shifts from deformylation to methionine cleavage. The RPBs moderately slows the PDF reaction, but significantly and selectively reduces the rates of the MAP reaction for substrates with larger second residue side chains. (B) RPBs define an optimal window for NME during translation. The relative abundance of individual ribosomal species at each nascent chain length was simulated during the cotranslational NME of FtsQ-V2 using the mathematical model and depicted using the indicated coloring scheme. PDF and MAP can act on the nascent proteins as soon as 45 residues are translated. PDF has an intrinsic preference for substrates of ∼70 residues in length (second row). The optimal window for the reaction of MAP is similar, but is imposed by the effects of RPBs (third row).