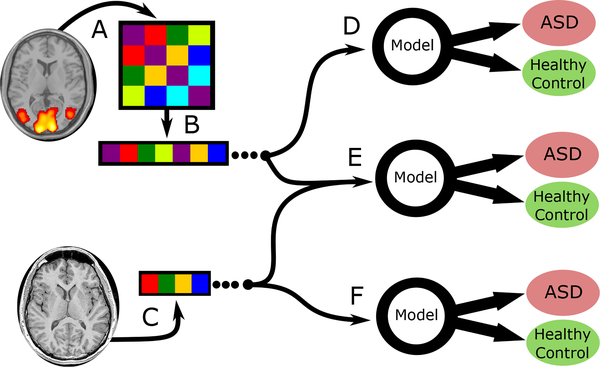
Fig. 1.
Combinations of derived features used by the predictive models tested in this study. Image data (rs-fMRI and sMRI) and was gathered externally by IMPAC organizers. (A) The rs-fMRI was transformed into a symmetric connectivity matrix for each atlas. (B) Upper triangular elements of matrix were flattened into a 1D vector. (C) The sMRI was transformed into a vector of cortical/subcortical ROI volumes and cortical thickness features. In (D) the connectivity matrix vector is used as the sole input for the predictive model, in (E) both anatomical and connectivity derived feature vectors are appended and used, while in (F) the anatomical features are used as the sole input for the predictive model.