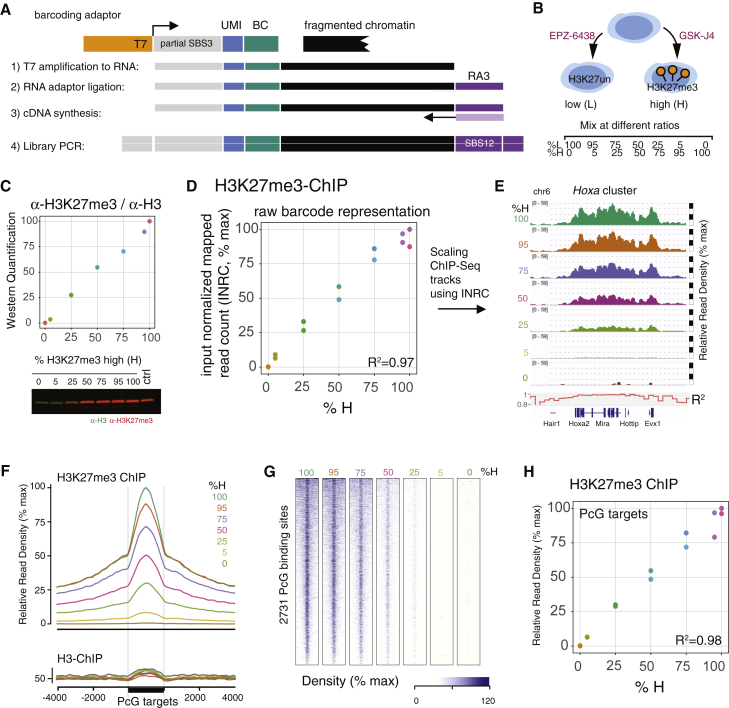
Figure 1.
MINUTE-ChIP Method Accurately Reflects Global and Local Proportions
(A) Schematic of MINUTE adaptor design for barcoding chromatin. One-sided ligation of adaptor comprising T7 RNA polymerase promoter (T7), random 6-bp sequence (UMI), and 8-bp barcode (BC) is sufficient for subsequent linear amplification. cDNA is primed from a ligated RNA adaptor (RA3). SBS3 and SBS12 designate standard Illumina read1 and read2 sequencing primers.
(B) Setup for calibration experiment: mESC treated with EZH2 inhibitor EPZ-6438 (resulting in depletion of H3K27me3, “L”) or JMJD3/UTX inhibitor GSK-J4 (resulting in very high H3K27me3, “H”) were mixed in defined ratios to establish a gradient of H3K27me3 with seven artificial conditions (0%, 5%, 25%, 50%, 75%, 95%, and 100% H).
(C) Quantitative infrared fluorescent western blot to confirm mixing ratios. Control sample (ctrl) is an untreated mESC in 2i condition. Raw western blot signal (α-H3K27me3 fluorescence over α-H3 fluorescence) was arbitrarily scaled between 0 and 100. Representative blot of three technical replicates is shown.
(D) MINUTE-ChIP quantification after α-H3K27me3 immunoprecipitation: input-normalized raw mapped read counts (INRCs) for two technical replicates with different barcodes are plotted against mixing ratio. INRC is subsequently used to quantitatively scale ChIP-seq tracks. Coefficient of determination (R2) for a linear regression is given.
(E) Example genomic regions showing quantitatively scaled H3K27me3 ChIP-seq tracks (average of both replicates) arising from different mixing ratios. Scale bar on right side breaks at 25%, 50%, 75%, and 100% maximal signal. Coefficient of determination (R2) was calculated over 10-kb windows.
(F) H3K27me3 signal (average of two replicates) across 2,731 polycomb (PcG) target sites. For comparison, H3 MINUTE-ChIP data are shown below. SE is rendered as shaded area around lines.
(G) Heatmap of the same data across 2,731 PcG target regions.
(H) Quantification of mean H3K27me3 signal at PcG target sites by replicate. Coefficient of determination (R2) for a linear regression is given.
See also Figure S1.