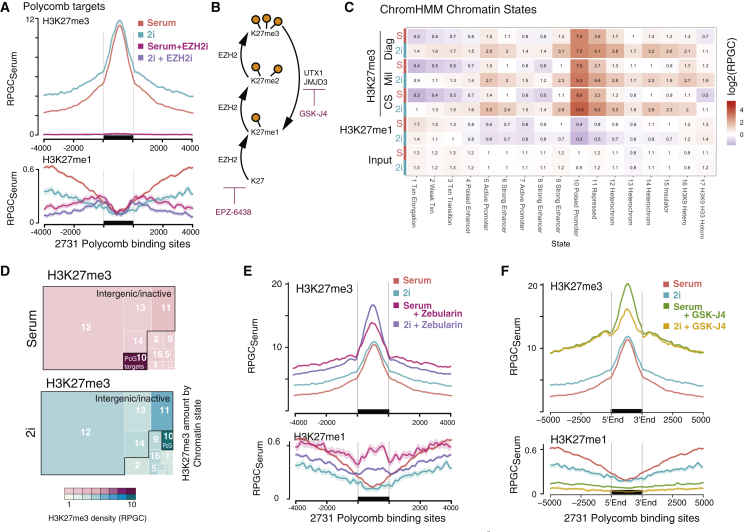
Figure 4.
H3K27me3 Is Maintained at PcG Targets and Gained Genome-wide
(A) Quantitative H3K27me3 and H3K27me1 profiles at 2,731 PcG target sites, in serum and 2i condition, as well as after treatment with EZH2 inhibitor EPZ-6438 (EZH2i) for 7 days. y axis shows reads per genomic content relative to serum condition (RPGCSerum), where serum condition is scaled to 1× coverage. SE is rendered as shaded area around lines.
(B) Schematic of enzymes involved in adding and removing H3K27 methylation and specific inhibitors used in pool 3.
(C) Heatmap of average H3K27me3, H3K27me1, and input read density across 17 functional chromatin states. Quantitative comparison between 2i and serum is shown using three different H3K27me3 antibodies. Color scale represents log2 of each pairwise 2i versus serum comparison.
(D) Treemap of total H3K27me3 amount (integral) across 17 functional chromatin states (as in C). Area is true to total proportions of H3K27me3. Color gradient shows average density (as in C).
(E) Average profile of H3K27me3 and H3K27me1 levels at PcG targets, in serum and 2i condition, as well as after treatment with DNMT inhibitor Zebularin. SE is rendered as shaded area around lines.
(F) Average profile of H3K27me3 and H3K27me1 levels at PcG targets, in serum and 2i condition, as well as after treatment with demethylase inhibitor GSK-J4.
See also Figures S3 and S4.