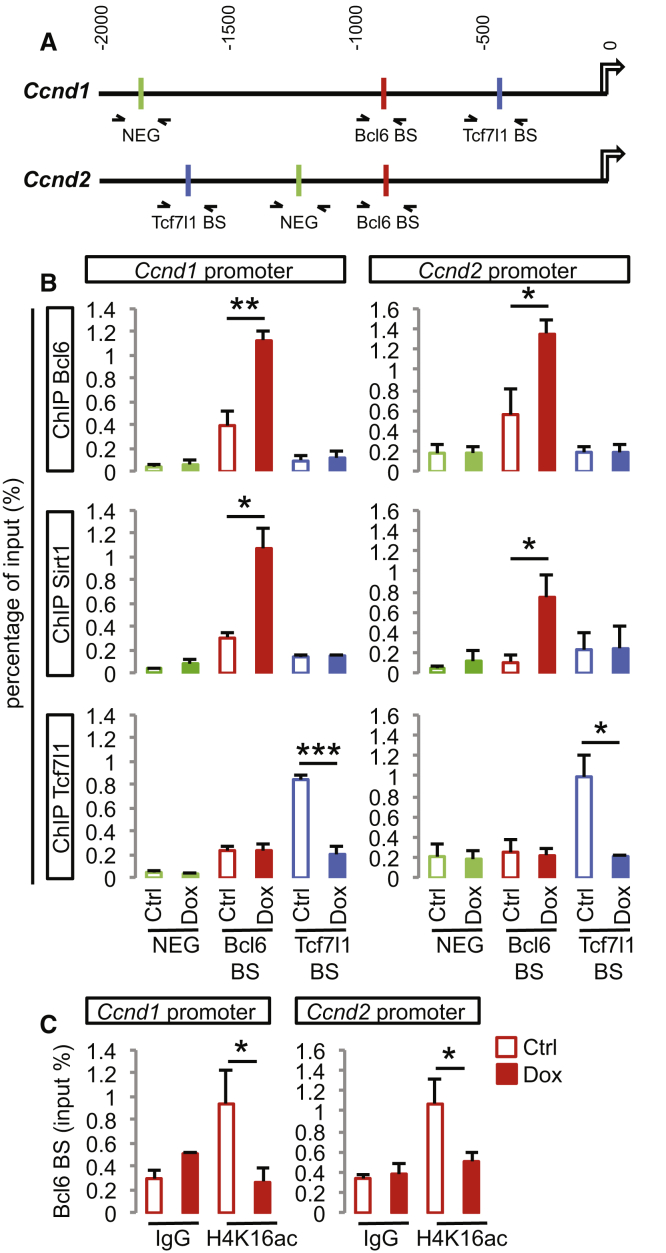
Figure 6.
Bcl6 and Sirt1 Bind to Ccnd1 and Ccnd2 Regulatory Regions, Leading to the Removal of the β-Catenin Effector Tcf7l1
(A) Schematic representation of the genomic region 2 kb upstream of Ccnd1 and Ccnd2 transcription starting sites showing validated Bcl6 (red) as well as putative Tcf7l1 (blue) binding sites and negative regions for either transcription factor (green) as predicted by the Jaspar software (http://jaspar.genereg.net). The arrows represent the amplified regions by qPCR used to measure the enrichment following ChIP.
(B) ChIP-qPCR analysis of the Bcl6, Sirt1, and Tcf7l1 binding sites on the Ccnd1 and Ccnd2 regulatory regions in cortical progenitors derived from Bcl6 A2 lox.Cre mouse ESCs (differentiation day 12, 24 h DMSO [Ctrl] or Dox treatment). Data are presented as mean + SEM of input enrichment (n = 3 differentiations). ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 using Student’s t test.
(C) ChIP-qPCR analysis of the H4K16ac histone acetylation mark on Bcl6 binding sites of the Ccnd1 and Ccnd2 regulatory regions in cortical progenitors derived from Bcl6 A2 lox.Cre mouse ESCs (differentiation day 12, 24 h DMSO [Ctrl] or Dox treatment). Data are presented as mean + SEM of input enrichment (n = 3 differentiations). ∗p < 0.05 using Student’s t test.
See also Figure S7.