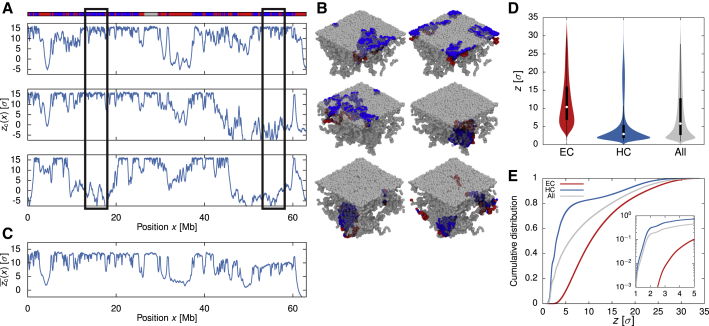
Figure 4.
Simulations Show Stochasticity in Chromatin Contacts with the NL
(A) Plots showing the distance of each chromatin bead from the horizontal plane at the center of the simulation box for three simulation runs in the growing (AC) phase . As the NL is located at the top of the box , a high value in indicates that the bead is close to the NL. The x axis shows the genomic position corresponding to each bead. The top track indicates the chromatin state along the polymer.
(B) Snapshots of the simulation runs coloring only the beads within the two highlighted regions in (A) (rectangular boxes). These figures reveal that the same chromatin segment can reside in different positions relative to the NL in different runs, which is consistent with experimental observations that LADs associate with the NL stochastically.
(C) The average value of for each bead over 20 simulations.
(D) Violin plots showing the distributions of the distance z from the NL for EC, HC, and all beads.
(E) Cumulative distributions of z for EC, HC, and all beads. The inset shows the distributions (in log-linear form) for small z.