Fig. 6.
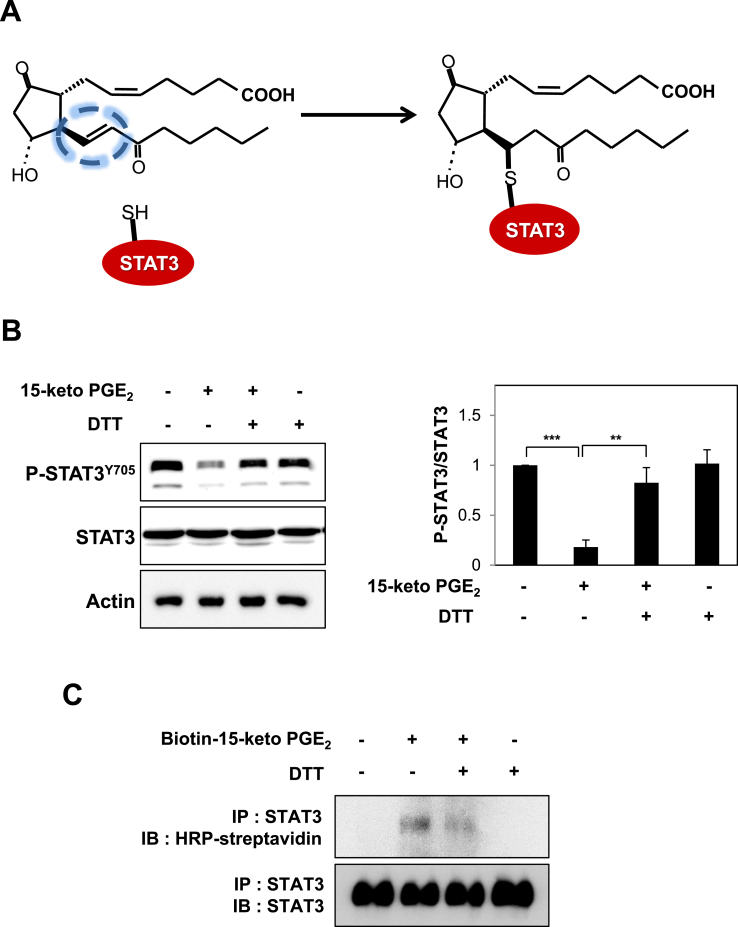
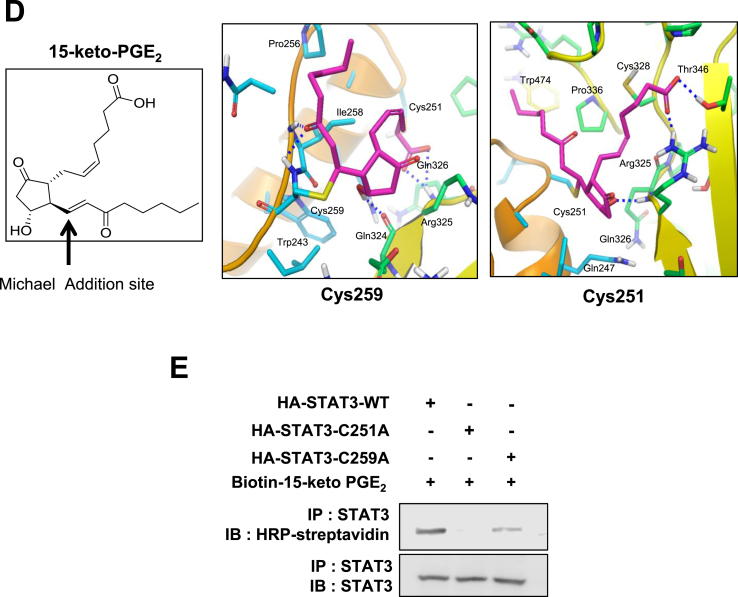
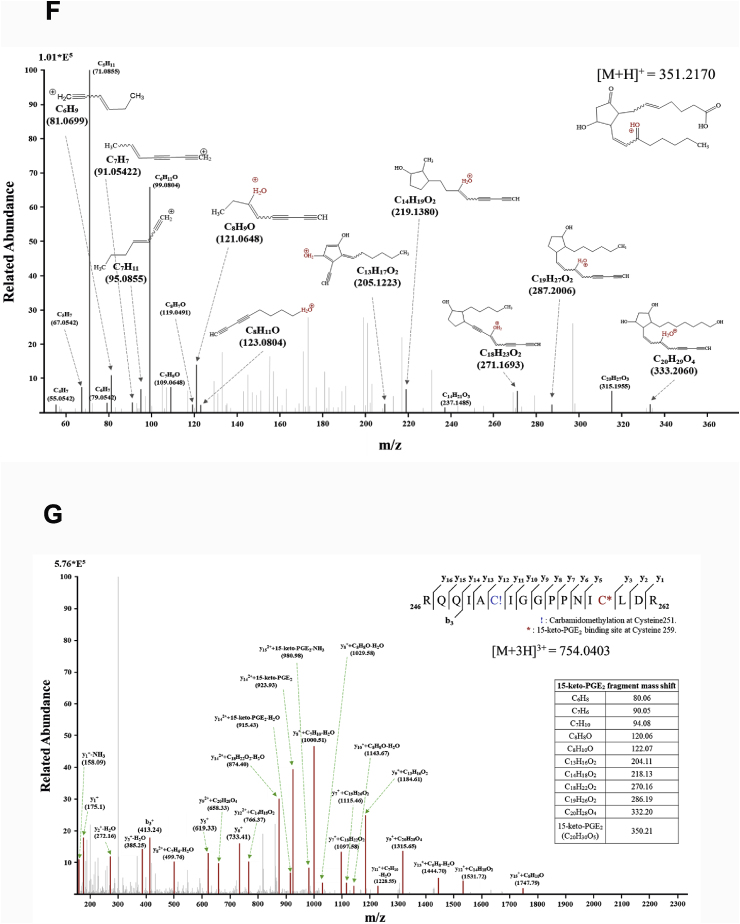
Covalent modification of STAT3 by15-keto PGE2. A. A proposed interaction between 15-keto PGE2 and a thiol group of STAT3. B. MCF10A-ras cells were pretreated with a thiol reducing agent, DTT (100 μM), for 1 h followed by exposure to 15-keto PGE2 (20 μM) for an additional 24 h. The protein levels of P-STAT3Y705 and STAT3 were determined by Western blot analysis. **p < 0.01, ***p < 0.001. C. The direct binding of 15-keto PGE2 to STAT3 was assessed using the avidin-biotin system. MCF10A-ras cells were pretreated with DTT (100 μM) for 1 h and then treated with biotinylated 15-keto PGE2 (40 μM) for an additional 12 h. The biotinylated 15-keto PGE2-STAT3 complex was detected by the immunoprecipitation and Western blot analyses. D. A putative model of 15-keto PGE2 covalently bound to Cys259 or Cys251 of STAT3 as predicted by a docking study. The blue dotted line represents a H-bond. E. PC3 cells were transfected with WT or Cys251 or Cys259-mutated STAT3 followed by treatment with 15-keto PGE2 (40 μM) for 24 h. WT STAT3 and STAT3 mutants were detected by anti-STAT3 antibody. F. Annotated fragment MS/MS spectrum of 15-keto PGE2 in positive ESI mode. The elucidation of fragment ion at m/z 333.2073, 315.1951 from the structure is given in the inset of the spectra. G. Mass spectrum of the STAT3 peptide (RQQIACIGGPPNICLDR) with 15-keto PGE2 binding. Annotated MS/MS spectrum illustrating the binding of 15-keto PGE2 to STAT3 protein at cysteine 259 residues was identified by LC-MS/MS. An exclamation mark indicates a carbamidomethylation on Cys251, and an asterisk indicates 15-keto PGE2 binding to Cys259. The precursor ion [M+3H]3 + for peptide with 15-keto PGE2 binding is m/z 754.0403. The 15-keto PGE2 fragment ions are inserted into the spectrum as a table. Each molecular formula present with b and y ions on the spectrum represents the 15-keto PGE2 fragment ion bound on Cys259. The sample preparation and other experimental details for mass spectral analysis are described in the Materials and methods. Sequence-informative fragmentation ions are summarized on the peptide sequence.