Figure 8.
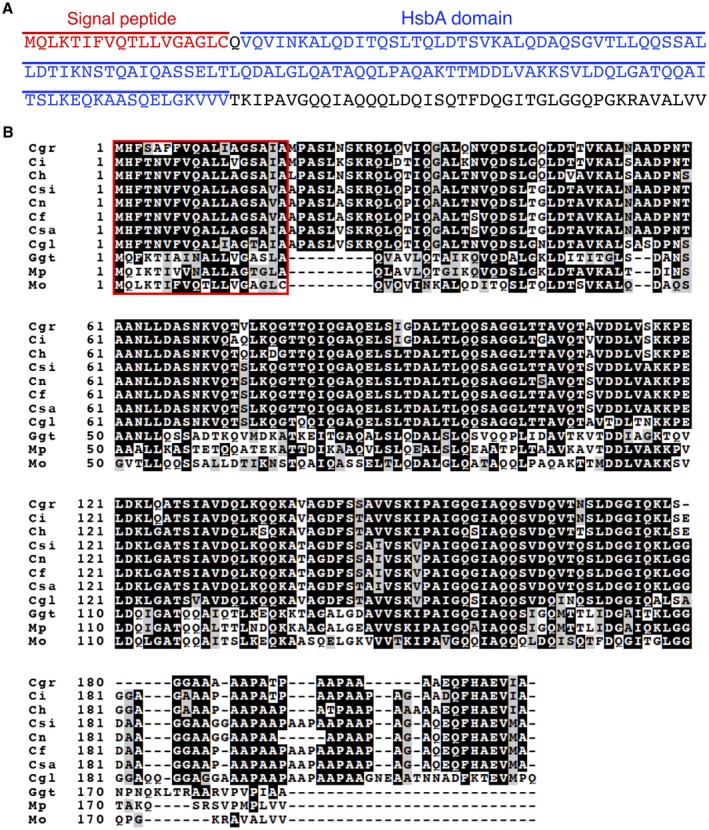
Amino acid sequence of MoSVP and sequence alignment with putative homologues from other fungal pathogens. (A) The predicted signal peptide and the HsbA domain of MoSVP are indicated in red and blue, respectively. (B) Amino acid sequence alignment of Magnaporthe oryzae MoSVP (Mo) with putative MoSVP homologues from Magnaporthiopsis poae (Mp), Gaeumannomyces graminis var. tritici (Ggt), Colletotrichum gloeosporioides (Cgl), C. salicis (Csa), C. fioriniae (Cf), C. nymphaeae (Cn), C. simmondsii (Csi), C. higginsianum (Ch), C. incanum (Ci) and C. graminicola (Cgr) generated using the ClustalW program (Tompson et al., 1994). Identical amino acids are indicated by white letters on a black background, similar residues are highlighted in grey and gaps introduced for alignment are indicated by hyphens. The putative signal peptide sequence is indicated by a red box.
