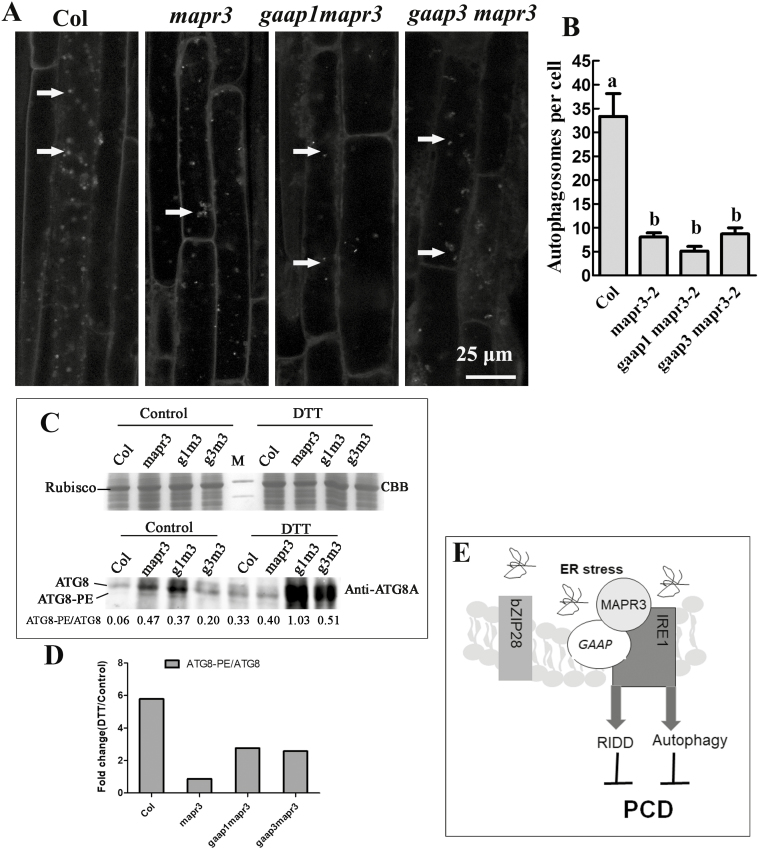
Fig. 9.
Mutations in MAPR3 and/or GAAP1/GAAP3 impaired the autophagy induced by ER stress. (A) Five-day-old seedlings of wild-type Col, mapr3-2, gaap1mapr3, or gaap3mapr3 were transferred to MS liquid medium supplemented with 4 mM DTT, followed by MDC staining. Then, roots were observed using confocal microscopy. Arrows indicate MDC-stained autophagosomes/autophagic bodies. Scale bar=25 μm. (B) The average number of MDC-stained autophagosomes per cortical cell in the root mature region was counted after DTT treatment as above. Data are average values ±SE (n=4) calculated from four independent experiments. For each experiment, >10 sections were used for the calculation for each genotype. Different lower case letters indicate significant differences (P<0.05 by two-way ANOVA and Tukey's range). (C) Total cellular proteins from the seedlings treated as in (A) were resolved on SDS–PAGE, stained with Coomasie brilliant blue (CBB) or probed with the ATG8A antibodies. The ratios of ATG8–PE to ATG8 in each line at the bottom were determined by ImageJ software. (D) The fold change of the ratio of ATG8–PE to ATG8 upon DTT treatment as in (C). (E) The working model in which GAAPs and MAPR3 are involved in regulating ER stress-induced autophagy and RNA decay by association with IRE1.