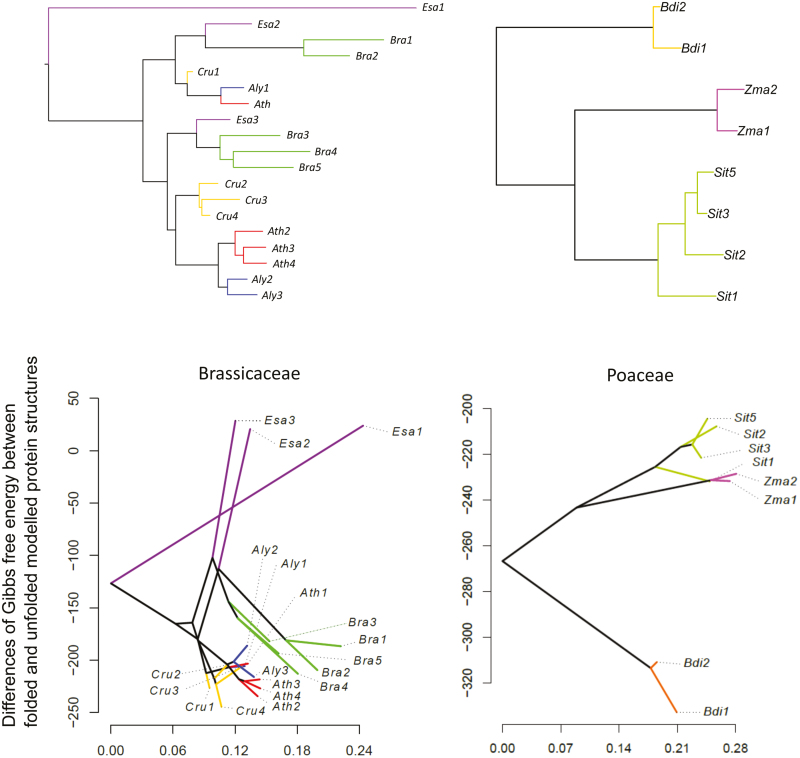
Fig. 5.
Stability of modelled RuBisCO structure. The phylogenetic trees of rbcS in Brassicaceae and Poaceae are shown in the upper panels. RuBisCO protein structures with RBCS encoded by each rbcS were estimated by homology modeling using Modeller (Eswar et al., 2008). The structure was repaired by the RepairPDB function of FoldX v4 (Schymkowitz et al., 2005). The stability of the whole RuBisCO was estimated using the ‘Stability’ function of FoldX4. Then, the result of protein stability was taken as a trait and phylogenetic relationships were given as input trees. We then drew a phenogram using the phytools package (Revell, 2012) in R (lower panels). Sit5 is shown as representative of Sit4/Sit5 because of synonymous substitutions.