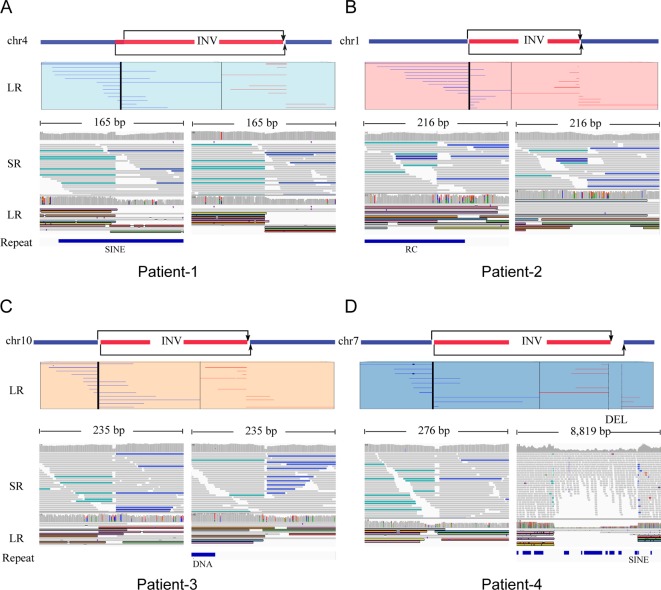
Figure 2.
Large inversion variants detected by Nanopore long-read sequencing technology. Four cases, patients 1–4, were represented by (A), (B), (C) and (D), respectively. For each case, the top panel figured the chromosome which inversion occurred supported by karyotype results, and long-read (LR) alignments coving the break points. Two parts of the split reads were denoted by blue (forward) and red (reverse). The panel in the bottom shows short-read alignments (BWA-MEM; IGV-SR) and long-read alignments (ngmlr; IGV-LR) around break points using Integrative Genomics View (IGV). All the break points, three of eight located in repeat region, were supported by short and long reads, except one in patient 4, DEL-INV-DEL, was missed by callers by using short reads. DEL, deletion; INV, inversion; RC, Rolling Circle; SINE, Short interspersed nuclear elements.