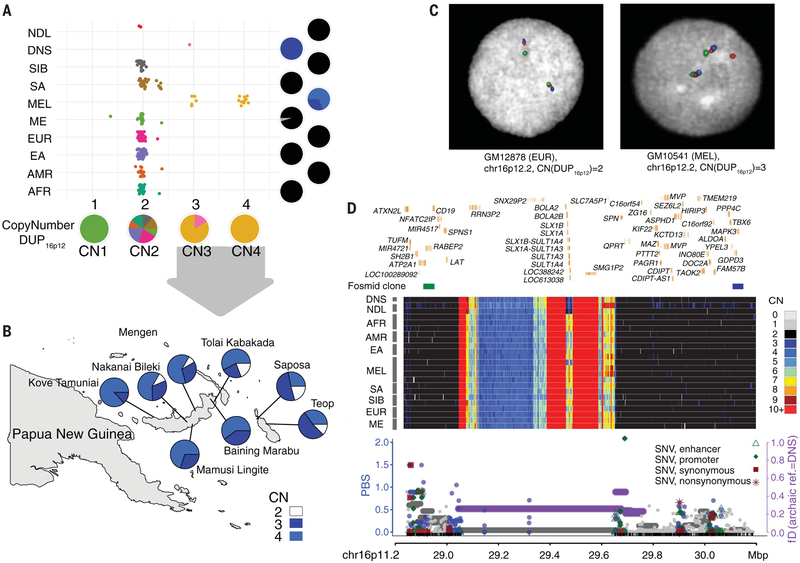
Fig. 2. Evidence for adaptive Denisovan introgression of the chromosome 16p12.2 duplication at 16p11.2 in Melanesians.
(A) Copy number (CN) estimates of DUP16p12 for the SGDP populations and three archaic samples. Pie charts indicate the CN frequency of populations (right) and the population fraction of CN genotypes (bottom). NDL, Neanderthals; DNS, Denisovans; SIB, Siberians; SA, South Asians; MEL, Melanesians; ME, Middle Easterners; EUR, Europeans; EA, East Asians; AMR, Native Americans; AFR, sub-Saharan Africans. (B) Geographic distribution for the DUP16p12 duplication genotypes of 242 independent blood-derived DNA samples from Melanesia. The CN color scheme matches that in (D). (C) FISH experiments using fosmid clones from 16p12.2 confirm an additional copy of DUP16p12 (red fosmid clone, 174222_ABC10_2_1_000044550500_M3 at 16p12.2; table S13; fig. S38) in a Melanesian cell line (GM10541, CN3) as opposed to a European cell line (GM12878, CN2). (D) (Top) Signals of adaptive introgression in the Melanesians at 16p11.2—the locus in which the DUP16p12 duplication was inserted. The heat map shows the CN distribution at chromosome 16p11.2. Fosmid clones (green: ABC10_000044688200_G16; blue: ABC10_000043626100_E12; table S13) indicate the region where the integration of DUP16p12 occurred at the 16p11.2 locus. (Bottom) PBS (left y axis) for SNVs (dots) and fD [horizontal lines, representing windows of 100 SNVs, computed using Denisovans as the archaic reference, right axis] at DUP16p12. Colored circles (blue) and/or horizontal lines (purple) indicate significant test statistics (P < 0.05). Note that introgression signals at both 16p12.2 and 16p11.2 disappear if Neanderthals are used as the archaic reference in the fD computation (figs. S36 and S47).