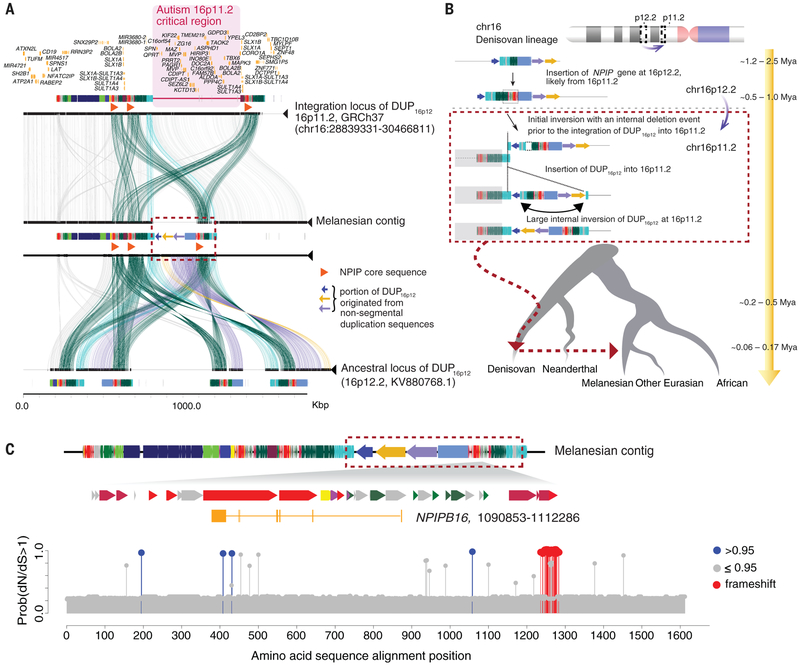
Fig. 3. Reconstruction of the structure and evolutionary history for the Melanesian–Denisovan duplication at chromosome 16p11.2.
(A) Structural comparison of chromosome 16p11.2 (human genome reference GRCh37; top), the structure-resolved Melanesian contig (middle) at 16p11.2, and the ancestral locus of DUP16p12 at 16p12.2 (KV880768.1; NCBI BioProject: PRJNA31257; bottom). Colored boxes denote annotated human segmental duplications, and lines connecting the sequences show regions of homology. Duplicated segments specific to the Melanesian genome (red dashed box) are indicated if derived from unique (colored arrows) or previous duplication (colored rectangles) sequences. The region of recurrent genome rearrangements associated with autism is highlighted (pink shaded area). (B) Schematic model for the evolution of the DUP16p12 duplication. The schematic depicts structural changes over time, leading to the Melanesian architecture. Evolutionary timing was estimated on the basis of a series of phylogenetic analyses using structure-resolved sequences from 16p12.2 and 16p11.2 loci (31). The absence of intermediate genomes makes the order of some structural changes uncertain. (C) A new member of the NPIPB gene family, NPIPB16 (1206 amino acids), in the Melanesian DUP16p12 sequence with predicted sites of positive selection. dN/dS analyses show positively selected amino acid substitutions at NPIPB16 lineages (blue circles) compared with other NPIPB genes. Note that the cluster of massive amino acid changes (red circles) at position 1236 to 1284 (alignment space) is predicted to result from two indel events in the C terminus of NPIPB16 as opposed to a series of independent amino acid substitution events (fig. S51).