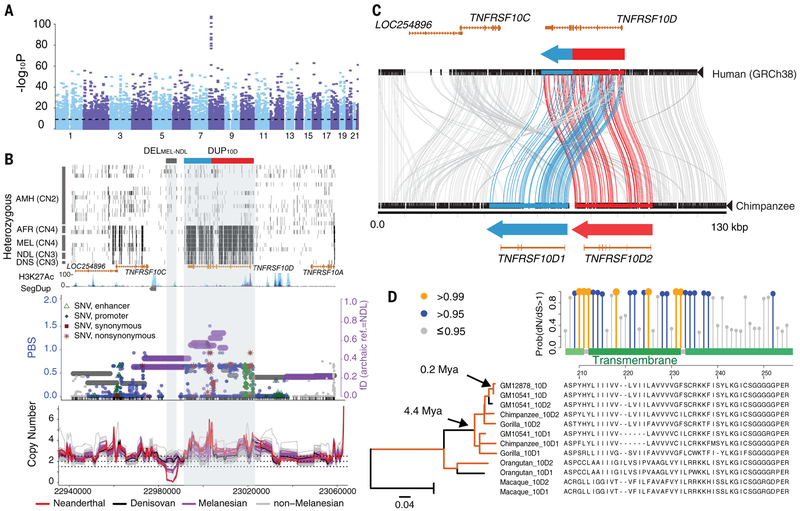
Fig. 4. Highly stratified CNVs at 8p21.3 in Melanesians and evidence for gene duplication and fusion followed by adaptive evolution at the TNFRSF10D locus.
(A) Manhattan plot for the P values of window-based FST test. The horizontal dashed line indicates the genome-wide Bonferroni-corrected significance. (B) (Top) Distribution of heterozygous sites (short black vertical bars) for a subset of the SGDP samples. The gray box at the top shows the location of DELMEL-NDL, whereas the blue-red box indicates the derived TNFRSF10D form, a fusion of TNFRSF10D1 (blue) and TNFRSF10D2 (red), as shown in Fig. 3C. (Bottom) Distributions of fD and PBS statistics, as well as CN trajectories of all samples across the region. (C) Comparison (Miropeats) of the major human allele versus chimpanzee genome structure, showing the tandem organization of the DUP10D variant and the predicted gene models. (D) Branch-site test of positive selection (dN/dS) using FLNC transcript data shows significant selection signals (P = 0.005) compared with the null model and a cluster of positively selected amino acid substitutions at the transmembrane domain of TNFRSF10D. Coding-sequence phylogeny shows significant positive selection (orange; dN/dS ratios >1; P < 0.05) on specific branches. Note that the orangutan paralogous sequences form a single clade as a result of interlocus gene conversion (fig. S60).