Abstract
Brain imaging genetics aims to reveal genetic effects on brain phenotypes, where most studies examine phenotypes defined on anatomical or functional regions of interest (ROIs) given their biologically meaningful annotation and modest dimensionality compared with voxel-wise approaches. Typical ROI-level measures used in these studies are summary statistics from voxel-wise measures in the region, without making full use of individual voxel signals. In this paper, we propose a flexible and powerful framework for mining regional imaging genetic associations via voxel-wise enrichment analysis, which embraces the collective effect of weak voxel-level signals within an ROI. We demonstrate our method on an imaging genetic analysis using data from the Alzheimers Disease Neuroimaging Initiative, where we assess the collective regional genetic effects of voxel-wise FDGPET measures between 116 ROIs and 19 AD candidate SNPs. Compared with traditional ROI-wise and voxel-wise approaches, our method identified 102 additional significant associations, some of which were further supported by evidences in brain tissue-specific expression analysis. This demonstrates the promise of the proposed method as a flexible and powerful framework for exploring imaging genetic effects on the brain.
Keywords: imaging genetics, enrichment analysis, genetic association analysis, voxel-wise analysis
I. INTRODUCTION
Imaging genetics is an emerging research field investigating the influence of genetic variants such as single-nucleotide polymorphisms (SNPs) on imaging phenotypes. Brain imaging genetics aims to reveal associations between genetic variations and quantitative traits (QTs) extracted from brain imaging data. These imaging QTs (iQTs) are measures extracted from either a single voxel [1] or a region of interest (ROI) [2]–[4] in the brain. An ROI is a pre-defined brain area containing a cluster of voxels with the same anatomical or functional annotation. Of note, the number of ROIs is much smaller than the number of voxels in the brain. Thus, most studies examine ROI-level phenotypes due to (1) modest dimensionality compared with voxel-wise approaches for increased statistical power, and (2) biologically meaningful annotation for easy interpretation.
Most existing ROI-level imaging genetic studies evaluate the associations between individual SNPs and ROI-level iQTs which are often defined as summary statistics (e.g., mean) of all the voxel-wise measures in the ROI. For example, genome-wide association studies (GWAS) have been performed for these iQTs and have discovered genes susceptible to various brain ROIs [5]–[7]. Targeted genetic studies have also been performed on brain ROIs to relate candidate SNPs to brain regions. However, most ROI-based approaches simply collapse voxel-level measures into a single value, and might lead to false-negative results when only weak signals exist in part of an ROI. Although voxel-wise strategies have been proposed to explore fine-grained variances of brain (e.g., [1], [8]), their effectiveness suffers from major multiple comparison issue due to ultra-high dimension of imaging and genetics data [8]. Cluster-wise approaches have been proposed to overcome the above limitation by identifying local voxel clusters to reach a pre-defined significant threshold [9]. The approach, however, ignores ROI-based anatomical or functional annotation.
Pathway enrichment analysis is a widely used method in genetics, where gene sets corresponding to biological pathways are examined for association with a phenotype to help increase statistical power and improve biological interpretation. Numerous studies on complex diseases have demonstrated that genes functioning in the same pathway can influence iQTs collectively even when constituent SNPs do not show significant association individually [3]. With these observations, in this work, we introduce enrichment analysis into imaging domain and propose an enrichment-based ROI-level imaging genetic association study (eIGAS) framework that estimates the collective genetic association with all the voxels in an ROI. To show the effectiveness of the eIGAS framework, we compare it with traditional ROI-based and voxel-based approaches via an imaging genetic study in Alzheimer’s disease (AD). Because the computational cost of voxel-wise GWAS is extremely expensive, we test our method in a targeted analysis between 19 AD candidate SNPs and brain-wide imaging phenotypes in 116 ROIs. We demonstrate that the proposed method outperforms the other two strategies with improved statistical power and biological interpretability.
II. MATERIALS AND METHODS
To demonstrate the power of the proposed eIGAS framework, we apply it to an FDG-PET imaging genetic analysis in AD. FDG-PET has been used to measure cerebral metabolic rates of glucose (CMRglc), and its change occurs early in AD.
A. Imaging and genotyping data
The imaging and genotyping data used in this article were obtained from the ADNI database (adni.loni.usc.edu). Preprocessed FDG-PET scans were downloaded from the ADNI website, then aligned to each participant’s same visit scan and normalized to the Montreal Neurological Institute (MNI) space as 2×2×2 mm voxels. FDG measurements of 185,405 voxels were extracted, and 116 ROIs were further computed using the mean of voxels within each ROI based on the MarsBaR AAL atlas as described in [7]. The number of voxels within 116 ROIs ranges from 54 to 5,104. 998 non-Hispanic Caucasian participants (Table I) with complete baseline voxel-level and ROI-level FDG measurements were studied.
TABLE I:
Participant characteristics
| Subjects | HC | SMC | EMCI | LMCI | AD |
|---|---|---|---|---|---|
| Number | 236 | 91 | 289 | 200 | 182 |
| Gender(M/F) | 122/114 | 38/53 | 163/126 | 117/83 | 109/73 |
| Age(mean±sd) | 76.2±6.7 | 72.5±5.6 | 71.7±7.2 | 74.3±8.3 | 75.4±7.7 |
| Edu(mean±sd) | 16.4±2.7 | 16.9±2.6 | 16.1±2.6 | 16.3±2.8 | 15.9±2.7 |
| APOE ε4 | 26.81% | 32.97% | 43.06% | 50.50% | 64.84% |
HC=Healthy Control; SMC=Significant Memory Concern; EMCI=Early Mild Cognitive Complaint; LMCI=Late Mild Cognitive Complaint; AD=Alzheimer’s Disease.
Genotype data of both ADNI-1 and ADNI-GO/2 phases were downloaded, quality controlled, imputed and combined as described in [10]. 5,574,300 SNPs were obtained for all 998 subjects studied here. A list of 23 AD risk SNPs were analyzed, containing 21 SNPs from the large scale meta-analysis of AD [11] plus two well-known APOE SNPs (rs429358 and rs7412). Four SNPs were excluded as no imputed genotyping data available. In total, 19 AD risk SNPs were included in our imaging genetic analysis. Detailed information of the 19 studied SNPs are shown in Table II.
TABLE II:
Details of 19 AD risk SNPs.
| SNP | Chr | Position | Closest gene | Major/minor |
|---|---|---|---|---|
| rs6656401 | 1 | 207692049 | CR1 | G/A |
| rs6733839 | 2 | 127892810 | BIN1 | C/T |
| rs35349669 | 2 | 234068476 | INPP5D | C/T |
| rs190982 | 5 | 88223420 | MEF2C | A/G |
| rs10948363 | 6 | 47487762 | CD2AP | A/G |
| rs2718058 | 7 | 37841534 | NME8 | A/G |
| rs1476679 | 7 | 100004446 | ZCWPW1 | T/C |
| rs11771145 | 7 | 143110762 | EPHA1 | G/A |
| rs28834970 | 8 | 27195121 | PTK2B | T/C |
| rs9331896 | 8 | 27467686 | CLU | T/C |
| rs10838725 | 11 | 47557871 | CELF1 | T/C |
| rs983392 | 11 | 59923508 | MS4A6A | A/G |
| rs10792832 | 11 | 85867875 | PICALM | G/A |
| rs17125944 | 14 | 53400629 | FERMT2 | T/C |
| rs10498633 | 14 | 92926952 | SLC24A4-RIN3 | G/T |
| rs4147929 | 19 | 1063443 | ABCA7 | G/A |
| rs429358 | 19 | 45411941 | APOE | T/C |
| rs3865444 | 19 | 51727962 | CD33 | C/A |
| rs7274581 | 20 | 55018260 | CASS4 | T/C |
B. Targeted genetic association study of FDG-PET imaging
We performed targeted genetic analysis of FDG-PET imaging measures on each voxel and each ROI, using linear regression under an additive genetic model in PLINK [12], with age, gender and education as covariates. Post-hoc analysis used Bonferroni correction for adjusting both the number of SNPs and the number of iQTs (i.e., voxel number for voxel-level analysis and ROI number for ROI-level analysis).
For comparison purpose, we constructed a novel ROI-level P-value using a summarized statistic from the voxel-level P-values, borrowing the idea from gene set analysis which maps SNP-level P to gene-level P [13]. Here we chose the second-best voxel-level P-value in each ROI to represent the ROI-level P, to avoid spurious associations from the best P.
C. Enrichment-based IGAS (eIGAS) framework
Pathway enrichment analysis has been widely used in genomic domain to examine gene sets corresponding to biological pathways for association with phenotypes. In this paper, we consider brain ROIs as pathways, each of which contains a set of voxels; and aim to identify ROIs significantly enriched by voxel-level genetic findings to form ROI-level genetic associations. Below, we describe the proposed method.
We propose the enrichment-based imaging genetic association study (eIGAS) framework using the over-representation analysis (ORA). We obtain the voxel-wise genetic association results from Subsection II-B, including P-values between S = 19 AD SNPs and N = 185, 405 voxels. Given a SNP Si, the imaging genetic findings are a list Li of significant SNP-voxel associations with P values passing a pre-defined threshold. Given an ROI Rk that contains total rk voxels , eIGAS aims to determine whether the set of voxels within targeted ROI Vk is enriched in Li. Now we present our ORA-based eIGAS method. Given a SNP Si, we have N Si-voxel associations from voxel-wise imaging genetic association analysis, out of which ni = | Li |(the set Li) are significant ones with P-value passing a pre-defined threshold. Out of these, we have rk = |Vk | associations from ROI Rk, of which li significant ones are from Li. Applying Fisher’s exact test for independence, the enrichment P-value for the ROI Rk associated with SNP Si is as follows:
| (1) |
Here, P r(·) is the probability function.
D. Evaluation of eIGAS
We evaluated the statistical power of eIGAS on discovering imaging genetic associations by comparing it with both ROI-based and second-best voxel-based approaches. We also validated the novel SNP-ROI findings in brain tissue-specific expression quantitative trait loci (eQTL) analysis. Specifically, we used eQTL dataset available in BRAINEAC (http://www.braineac.org/), a web server for data from the UK Brain Expression Consortium (UKBEC) [14]. This dataset contains ten brain tissues from 134 neuropathologically normal subjects. We assessed the altered gene expression of identified SNPs from eIGAS in the corresponding brain tissues.
III. RESULTS
A. Targeted genetic associations of FDG-PET iQTs
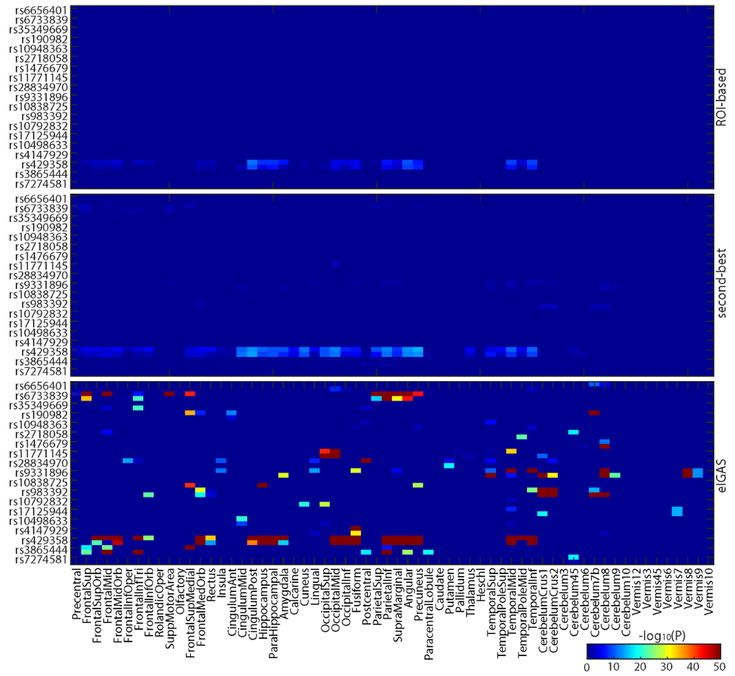
Targeted genetic analyses were performed on both ROI-level (i.e., mean of all voxels in the ROI) and voxel-level FDG measures, to examine imaging genetic associations between 19 AD SNPs and FDG measures from 116 ROIs and 185,405 voxels, respectively. To facilitate comparison among these methods, for the 185,405 voxel-level P results, we employed the second-best P-value strategy to map those to 116 ROI-level P summary statistics. Using Bonferroni corrected P < 0.05/(116×19) = 2.27e-5 as threshold, we identified 41 SNP-ROI hits from ROI-based approach covering 1 SNP (APOE rs429358) and 41 brain ROIs, and 91 SNP-ROI associations from the second-best strategy covering 6 SNPs and 78 brain ROIs. Detailed findings of these two strategies were shown in the top two panels of Fig. 1.
Fig. 1:
Comparison of brain ROI genetic analysis strategies. Shown from top to bottom are the results of traditional ROI-level analysis, traditional voxel-level analysis with the 2nd-best voxel representing the ROI, and the proposed eIGAS analysis.
B. Imaging genetic associations from enrichment-based IGEA
For each AD SNP, we obtained a list of 185, 405 SNP-voxel associations across all voxels in brain. Given a SNP Si, for each ROI, we assessed the collective effect of Si on all voxels within the ROI by calculating the enrichment score, to relate Si to ROI. We employed a relatively generous threshold 0.05/19 = 2.63e-3 to determine the list of significant SNP-voxel associations for eIGEA, to avoid missing individually moderate while collectively significant signals. We obtained enrichment P-values between 19 AD SNPs and 116 ROIs, among which 158 SNP-ROI pairs were significant after correcting for both the number of SNPs and the number of ROIs (i.e., P < 0.05/(116×19) = 2.27e-5). These eIGAS findings covered all 19 AD SNPs and 86 unique ROIs. Out of 158 findings, 102 SNP-ROI pairs were novel and 56 SNP-ROI pairs overlapped with findings from the prior two strategies.
Fig. 1 shows the results from eIGEA (bottom panel) and other two methods. As we expected, eIGAS not only conserved high concordance with findings from ROI-based and voxel-based second-best strategies, but also reported novel SNP-ROI associations. This indicates that the integration of fine-grained association statistics with brain ROI information would promote the identification of high-level imaging genetic associations and facilitate biological interpretation.
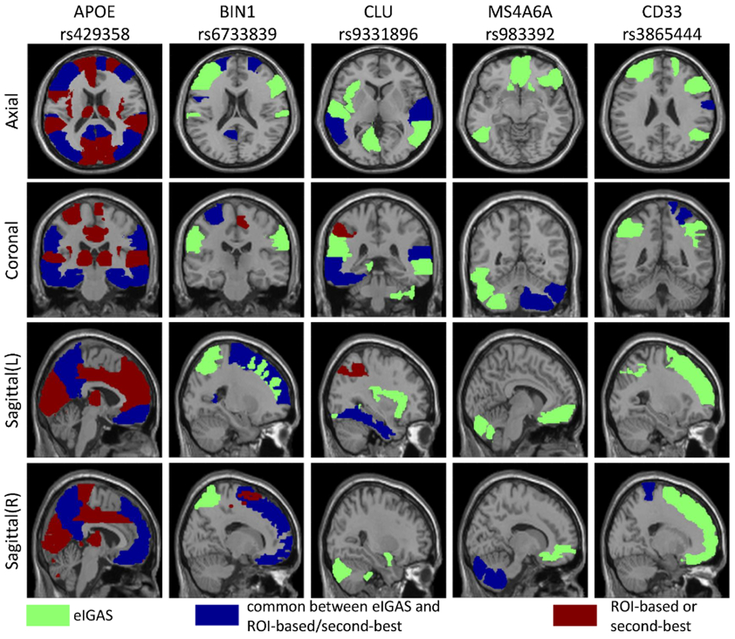
To better illustrate the findings from eIGAS as well as compare it with the other two strategies, we summarized eIGAS findings by ranking AD SNPs according to the number of their significantly related ROIs. Top five SNPs were extracted from eIGAS results, including rs429358, rs6733839, rs9331896, rs983392, rs3865444; and they were significantly associated with 39, 18, 17, 13, and 11 ROIs, respectively. Given these five SNPs, significantly associated ROIs from both ROI-based and voxel-based second-best approaches were also extracted.
Fig. 2 maps all these ROIs onto the brain, where ROIs were assigned different colors according to which approaches they were identified from. As the best-known AD risk variant, APOE rs429358 associated with the largest number of ROIs from three approaches, including various frontal, temporal, occipital, amygdala, precuneus and other regions responsible for different functions. Regarding the ROIs associated with other top SNPs, most of them were discovered by eIGAS only (green ones) or by both eIGAS and ROI-based and/or voxel-based second-best approaches (blue ones); while only few ROIs were reported by ROI-based and/or voxel-based second-best approaches (red ones). Given that disease risk variants can influence pathological behaviors through intermediate phenotypes, our studied SNPs might implicitly mediate FDG iQTs to contribute to AD. Thus our eIGAS framework promoted the identification of these intermediate traits for better understanding of the underlying disease mechanism.
Fig. 2:
Brain maps of ROIs associated with top eIGAS SNPs.
C. Biological significance of eIGAS findings
We further examine the biological significance of 102 new SNP-ROI findings identified from eIGAS, through brain tissue-specific eQTL analysis using genotyping and expression data of ten brain tissues from UK Brain Expression Consortium (UKBEC). There were totally 18 unique SNPs and 67 unique ROIs covered by 102 new hits. After mapping ROIs to UKBEC brain tissues, there remain 53 SNP-ROI pairs covering 16 SNPs and 34 ROIs. We assessed the effect of these 16 SNPs on brain tissue-specific expression levels of their nearest genes, and identified 15 significant tissue-specific eQTLs with P-value less than 0.05 (see Table III for details). This indicates the power of our method for identifying biologically meaningful imaging genetic associations.
TABLE III:
eQTL analysis of eIGAS findings.
| SNP | Chr | Gene | P | Tissue |
|---|---|---|---|---|
| rs10792832 | 11 | PICALM | 1.8e-2 | Occipital cortex |
| rs11771145 | 7 | EPHA1 | 1.4e-3 | Occipital cortex |
| rs11771145 | 7 | EPHA1 | 2.0e-3 | Thalamus |
| rs11771145 | 7 | EPHA1 | 1.8e-2 | Temporal cortex |
| rs28834970 | 8 | PTK2B | 3.8e-2 | Frontal cortex |
| rs28834970 | 8 | PTK2B | 1.1e-4 | Temporal cortex |
| rs28834970 | 8 | PTK2B | 6.0e-3 | Putamen |
| rs35349669 | 2 | INPP5D | 9.1e-3 | Frontal cortex |
| rs3865444 | 19 | CD33 | 3.3e-2 | Frontal cortex |
| rs4147929 | 19 | ABCA7 | 2.4e-2 | Temporal cortex |
| rs6656401 | 1 | CR1,CR1L | 1.9e-2 | Occipital cortex |
| rs6656401 | 1 | CR1,CR1L | 2.8e-3 | Temporal cortex |
| rs6733839 | 2 | BIN1 | 3.6e-2 | Frontal cortex |
| rs9331896 | 8 | CLU | 6.1e-5 | Temporal cortex |
| rs983392 | 11 | MS4A6A,MS4A4E | 1.1e-2 | Frontal cortex |
IV. DISCUSSION
In this paper, we have presented an enrichment-based imaging genetic association study (eIGAS) framework to explore the collective effect of a genetic variant on a brain region by integrating fine-grained voxel-wise imaging genetic associations with anatomically or functionally annotated ROI information. We have demonstrated its effectiveness using imaging genetics data from an AD study. In addition to associations identified by traditional ROI-based or voxel-based approaches, our approach has reported novel SNP-ROI findings and demonstrated their biological significance. This indicates the increased power of the presented method on identifying individually modest while collectively substantial signals.
The real power of eIGAS, however, can be affected by several factors. First, Fisher’s test requires a pre-defined threshold to determine the list of significant SNP-voxel pairs. Although this makes the framework more flexible in practice for tightening or relaxing voxel-level effects, it considers only the count of significant pairs without taking the full spectrum of association statistics. Rank-based enrichment strategies (e.g., [15]) can be employed in our framework to overcome these limitations. Another issue is that eIGAS requires to compute voxel-level associations in advance, which is both time and space demanding, especially given millions of SNPs in GWAS data. Therefore, another direction is to design parallel computational framework for accelerating the voxel-level GWAS. Another interesting future direction is to compare the performances between our enrichment-based approach and random field theory strategies as implemented in SPM (www.fil.ion.ucl.ac.uk/spm/).
Acknowledgments
This work was supported by NIH R01 EB022574, R01 LM011360, U19 AG024904, R01 AG019771, and P30 AG010133; and NSF IIS 1837964. The complete ADNI Acknowledgement is available at http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
Contributor Information
Xiaohui Yao, Email: Xiaohui.Yao@pennmedicine.upenn.edu, Biostatistics, Epidemiology and Informatics University of Pennsylvania, Philadelphia, PA.
Shan Cong, Email: congs@purdue.edu, Electrical and Computer Engineering Purdue University, West Lafayette, IN.
Jingwen Yan, Email: jingyan@iupui.edu, Informatics and Computing Indiana University, Indianapolis, IN.
Shannon L. Risacher, Email: srisache@iupui.edu.
Andrew J. Saykin, Email: asaykin@iupui.edu.
Jason H. Moore, Email: jhmoore@upenn.edu.
Li Shen, Email: Li.Shen@pennmedicine.upenn.edu.
REFERENCES
- [1].Stein JL, Hua X et al. , “Voxelwise genome-wide association study (vgwas),” NeuroImage, vol. 53, no. 3, pp. 1160–1174, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Risacher SL, Shen L et al. , “Longitudinal mri atrophy biomarkers: Relationship to conversion in the adni cohort,” Neurobiology of Aging, vol. 31, no. 8, pp. 1401–1418, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Yao X, Yan J et al. , “Two-dimensional enrichment analysis for mining high-level imaging genetic associations,” Brain Informatics, vol. 4, no. 1, pp. 27–37, March 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Shen L, Kim S et al. , “Whole genome association study of brain-wide imaging phenotypes for identifying quantitative trait loci in MCI and AD: A study of the ADNI cohort,” NeuroImage, vol. 53, no. 3, pp. 1051–1063, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Saykin AJ, Shen L et al. , “Genetic studies of quantitative MCI and AD phenotypes in ADNI: Progress, opportunities, and plans,” Alzheimers Dement, vol. 11, no. 7, pp. 792–814, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Shen L, Thompson PM et al. , “Genetic analysis of quantitative phenotypes in AD and MCI: imaging, cognition and biomarkers,” Brain Imaging and Behavior, vol. 8, no. 2, pp. 183–207, June 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Yao X, Yan J et al. , “Tissue-specific network-based genome wide study of amygdala imaging phenotypes to identify functional interaction modules,” Bioinformatics, vol. 33, no. 20, pp. 3250–3257, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hibar DP, Stein JL et al. , “Voxelwise gene-wide association study (vgenewas): Multivariate gene-based association testing in 731 elderly subjects,” NeuroImage, vol. 56, no. 4, pp. 1875–1891, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ge T, Feng J et al. , “Increasing power for voxel-wise genome-wide association studies: The random field theory, least square kernel machines and fast permutation procedures,” NeuroImage, vol. 63, no. 2, pp. 858–873, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kim S, Swaminathan S et al. , “Influence of genetic variation on plasma protein levels in older adults using a multi-analyte panel,” PLoS ONE, vol. 8, no. 7, p. e70269, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lambert JC et al. , “Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease,” Nat. Genet, vol. 45, no. 12, pp. 1452–1458, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Purcell S et al. , “PLINK: a tool set for whole-genome association and population-based linkage analyses,” Am. J. Hum. Genet, vol. 81, no. 3, pp. 559–75, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Nam D et al. , “GSA-SNP: a general approach for gene set analysis of polymorphisms,” Nucleic Acids Res, vol. 38, pp. W749–54, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ramasamy A, Trabzuni D et al. , “Genetic variability in the regulation of gene expression in ten regions of the human brain,” Nature Neuro-science, vol. 17, pp. 1418–1428, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Subramanian A, Tamayo P et al. , “Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles,” PNAS, vol. 102, no. 43, pp. 15 545–15 550, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]