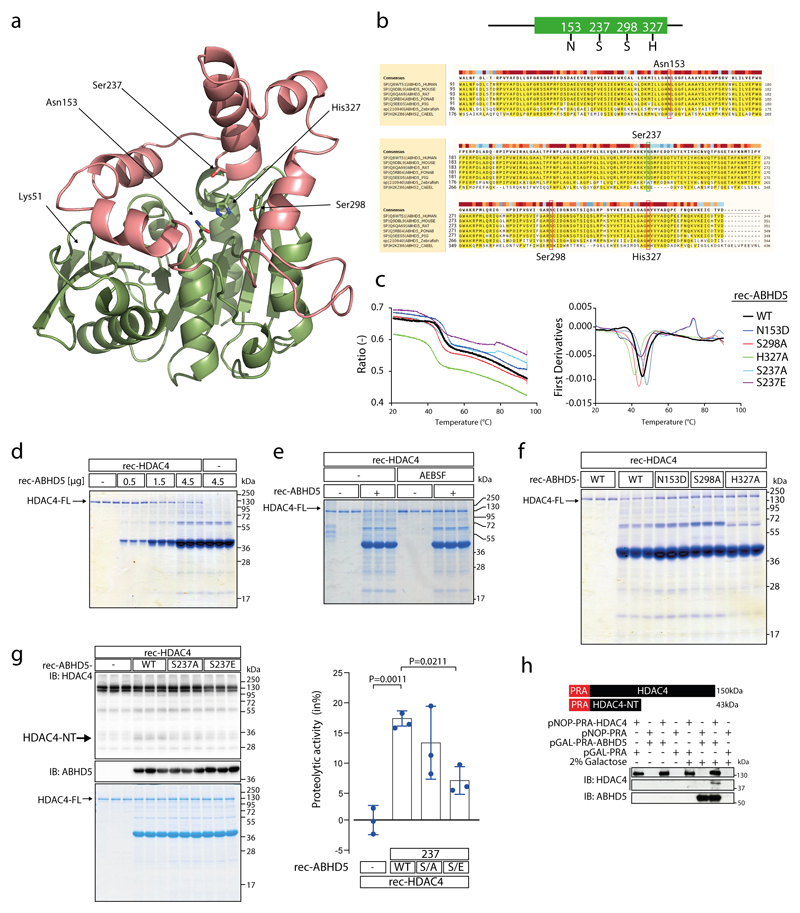
Extended Data Fig. 2. ABHD5 possesses intrinsic serine protease activity.
(a) Prediction of 3D structure of ABHD5 using SWISS-MODEL. Highlighted are amino acids (aa) predicted to serve as putative catalytic triad (Asn153, Ser298 and His327) and the putative phosphorylation site Ser-237. Ser-237 and Ser-298 of ABHD5 lie in spatial proximity to each other. Lys51 represents the N-terminal starting aa of the peptide sequence used for modeling. Pink: cap domain; green: a/b hydrolase core. (b) Multiple sequence alignment of ABHD5 protein of different species as indicated by UniProt online tool (uniprot.org/align/). Red boxes denote aa N153, S298 and H327 of the predicted serine protease catalytic triad, green box denotes aa S237 as reported PKA phosphorylation site (aa numbering corresponds to human ABHD5 protein). (c) Tryptophan fluorescence decay plot upon thermal denaturation of rec-ABHD5-WT, -N153D, -S237A, -S237E -S298A and -H327A (1°C/min). Left: the ratio of fluorescence at 350/330 nm was plotted against the temperature. Right: Equivalent analysis to identify the melting temperature (Tm) of the rec-proteins. Measurements were performed once. (d) Coomassie staining after protease assay using recombinant purified HDAC4 (rec-HDAC4) and increasing amounts of recombinant purified ABHD5 (rec-ABHD5) as indicated. (e) Coomassie staining after protease assay using rec-HDAC4 and rec-ABHD5 in the presence or absence of serine protease inhibitor AEBSF as indicated. (f) Coomassie staining after protease assay using rec-HDAC4 and rec-ABHD5 wild type (WT) or putative catalytic triad mutants as indicated. The data in (d-f) represent results of three independent experiments. (g) Immunoblotting and Coomassie staining for HDAC4 and ABHD5 after in vitro proteolysis assays involving Rec-HDAC4, rec-ABHD5 wild type (WT) or Ser-237 phospho-death (S237A) and phospho-mimetic (S237E) mutants as indicated. Coomassie staining was used to quantify the proteolytic activity of ABHD5. Statistical analysis: Values are presented as Mean±SEM, n=3 (n represents independent experiments); by one-way ANOVA, P<0.05 considered as significant. (h) Immunoblotting for HDAC4 and ABHD5 in yeast cell extracts after proteolysis assay in a yeast expression system as indicated. Because HDAC4 contains the Protein-A (PRA; 15 kDa) tag, HDAC4-NT is expected to migrate at 43 kDa. The experiment was performed at least three times with similar results.