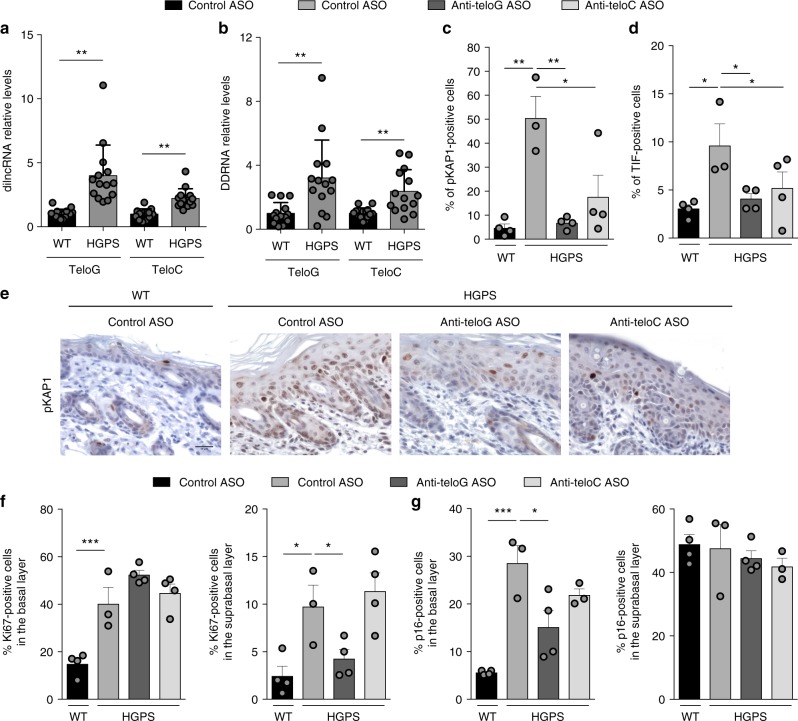
Fig. 3.
A mouse skin model of HGPS shows increased levels of tncRNAs and their inhibition in vivo reduces DDR activation and cellular senescence. a, b Total cell RNA was isolated from the skin of wild type (WT) and HGPS mice at post natal days 3 to 8. a tdilncRNAs were quantified by strand-specific RT-qPCR. Error bars represent the s.d. n = 14 mice per group. **P < 0.01; two-tailed Student’s t test. b tDDRNAs were quantified by miScript PCR amplification of gel-extracted small RNAs (shorter than 40 nucleotides). Error bars represent the s.d. n = 14 mice per group. **P < 0.01; two-tailed Student’s t test. c–g Mice subjected to systemic delivery of the indicated ASOs were sacrificed at post natal day 6 and skin samples were stained for DDR and proliferation markers. Mouse skin sections were immunohistochemically stained for pKAP1 (c, e) and positive cells were quantified in the epidermis. Scale bar, 50 μm. d Mouse skin sections were stained for pKAP1 and TRF1 to quantify telomere dysfunction-induced foci (TIFs) as determined by pKAP1 co-localizing with TRF1 in the basal layer of the skin. A cell was counted as positive if showing at least one TIF. n = 3 independent experiments. **P < 0.01; one-way ANOVA with multiple-comparison post-hoc corrections. At least 300 cells per sample were analyzed. Quantification of Ki67 (f) and p16 (g) positive cells in the supra basal and basal layers of epidermis. Error bars represent the s.d. n = 3–4 mice per group. *P < 0.05, **P < 0.01, ***P < 0.001; one-way ANOVA with multiple-comparison post-hoc corrections. At least 300 cells per sample were analyzed. Source data are provided as a Source Data file