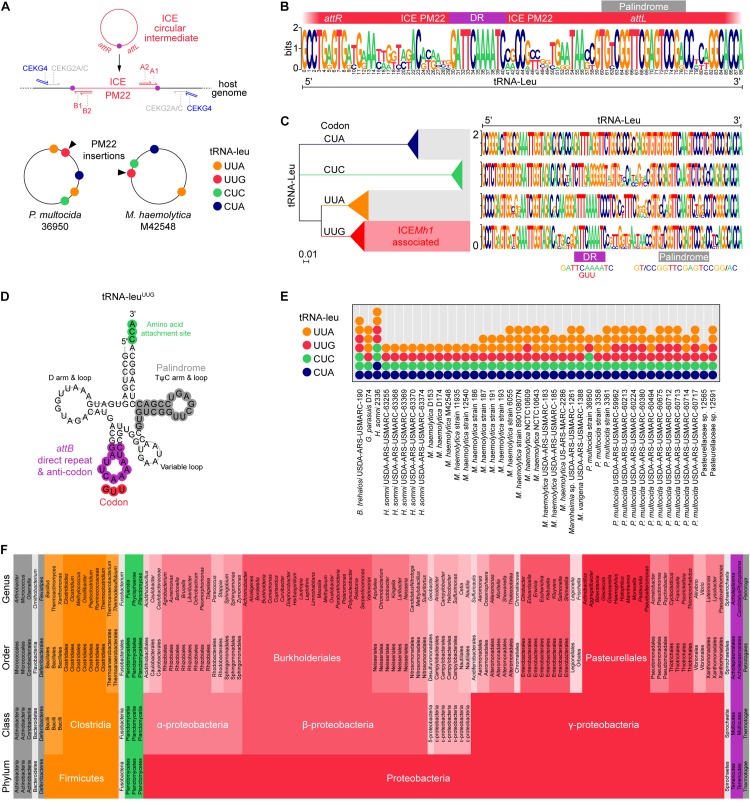
FIGURE 2.
Identification of tRNA-leuUUG as the putative attB insertion site of ICEMh1-like elements and predicted host range. (A) Schematic for ICE orientation and random PCR mapping of ICEMh1PM22 junctions in 18 Mannheimia haemolytica and 18 Pasteurella multocida transconjugants. ICEMh1PM22 insertions in every transconjugant were identical and aligned to a specific tRNA-leu in the genomes of P. multocida strain 36950 and M. haemolytica M42548. (B) Sequence logo of left and right MUSCLE-aligned junctions identified in 41 Pasteurellaceae harboring ICEMh1-like sequences. The left and right ends of ICEMh1-like variants contain a conserved direct repeat (DR; 5′-GATTCAAAATC-3′) attL conserves the tRNA TψC loop’s imperfect palindrome (5′-CGGTTCGAGTCCG-3′). The attL and attR sites are designated with respect to ICEPmu1 in Michael et al. (2011b). (C) Neighbor-joining tree of all tRNA-leu from 41 Pasteurellaceae harboring ICEMh1-like sequences. All ICEMh1-like insertions were associated with tRNA-leuUUG. (D) Predicted structure of tRNA-leuUUG showing presumptive ICEMh1 attB attachment site (also encoding the tRNA anticodon) and palindrome. (E) Frequencies of tRNA-leu codon types in 41 Pasteurellaceae harboring ICEMh1-like sequences. (F) Predicted host range of ICEMh1-like elements based on tRNA-leuUUG alignment by BLAST and strict (100% identity) conservation of direct repeat and palindrome. Colors are arbitrary.