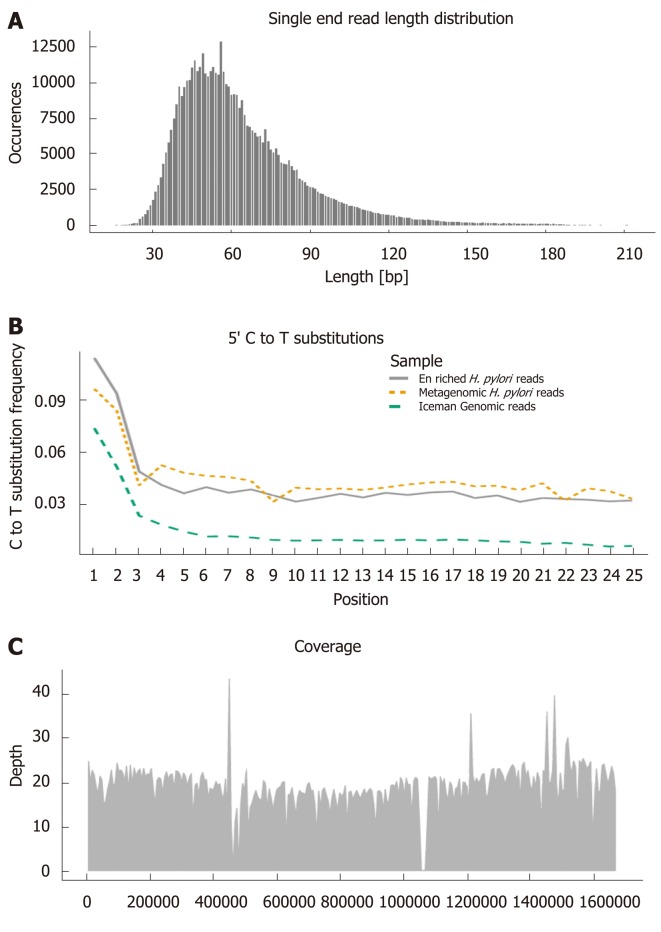
Figure 2.
Ancient DNA characteristics used for authenticating endogenous sequences. Visualised with Helicobacter pylori (H. pylori) sequences enriched from the gut of the Iceman and aligned to the H. pylori 26695 reference genome[20]: A: Read length distribution. B: C to T substitutions towards the 5’ end of sequence reads (Enriched H. pylori reads, metagenomic H. pylori reads, human Iceman genomic reads). C: Mean genome coverage and distribution of the aligned sequences. H. pylori: Helicobacter pylori.