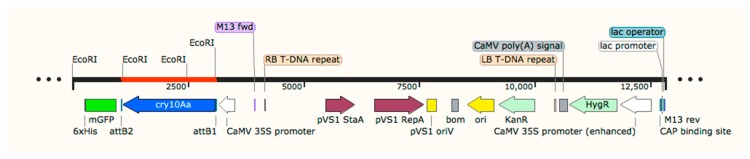
Figure 1.
Schematic representation of the plasmid pMDC85/cry10Aa. The 12,838 bp-long plasmid consists of the cry10Aa gene under 2XCaMV35S (cauliflower mosaic virus 35S promoter with a duplicated enhancer region) and 3′NOS (nopaline synthase terminator); HygR (hygromycin phosphotransferase) gene (hph) under 2XCaMV35S and 3′NOS terminator, mGFP (modified green fluorescent protein) under 2XCaMV35S promoter and 3′NOS terminator. 6xHis (6x histidine affinity tag); attB1-B2 (mutant version of attB); RB (right border repeat from nopaline C58T-DNA); pVS1 StaA (stability protein from Pseudomonas plasmid pVS1); pVS1 RepA (replication protein from Pseudomonas plasmid pVS1); Bom (bases of mobility region from pBR322); Ori (high-copy-number ColE1/pMB1/pBR322/pUC origin of replication); KanR (aminoglycoside phosphotransferase aphA-3, which confers kanamycin resistance); LB T-DNA repeat (left border repeat from nopaline C58 T-DNA); CaMV poly(A) signal (cauliflower mosaic virus polyadenylation signal); CAP binding site (E. coli catabolite activator protein); lac promoter (three segments) promoter for the E. coli lac operon; lac operator (laccase repressor encoded by lacI); and M13 rev. (common sequencing primer).