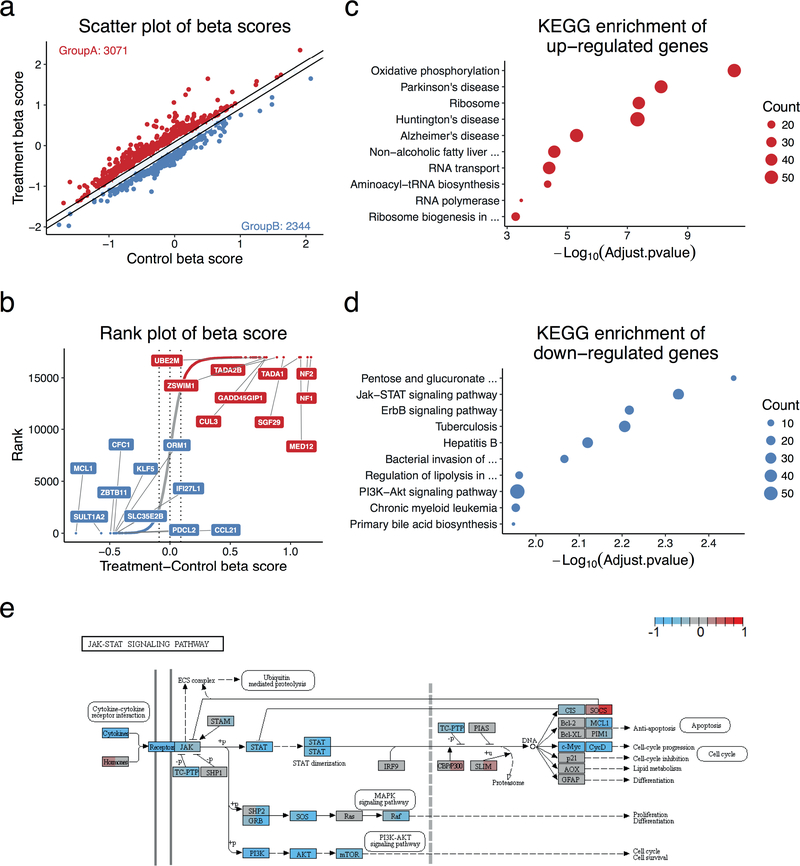
Figure 4. CRISPR/Cas9 screen analysis by MAGeCKFlute.
The data analysed here is a CRISPR screen in a breast melanoma cancer cell line, A375, treated with the BRAF protein kinase inhibitor vemurafenib (PLX)7. Data was processed with FluteMLE. (a) Scatterplot of treatment and control beta scores. The beta scores were normalized using the median of the beta scores of the core essential genes we compiled (Supplementary Data 2, Supplementary Method). The two diagonal lines indicate +/−1 standard deviation of the difference between treatment and control beta scores. Red dots (Group A) are genes whose beta score increased after treatment. Blue dots (Group B) are genes whose beta score decreased after treatment. (b) The genes are sorted based on the differential beta score, which is calculated by subtracting the control beta score from the treatment beta score. The colour scheme is the same as in panel a and dots between two diagonal lines are genes for which the beta score did not change significantly between different conditions. The top 10 enriched KEGG pathways with (c) positively (Red, Group A) and (d) negatively (Blue, Group B) selected genes. The p-value was calculated with the clusterProfile package that is based on the hypergeometric distribution. The size of each circle indicates the number of genes which are enriched in the corresponding function. (e) A visualization of treatment and control beta scores over the JAK-STAT signaling pathway generated by the Pathview package47. The left and right portion of a gene-box represent control and treatment beta scores, respectively. Red indicates a positive beta score, blue indicates a negative beta score, and grey marks genes are neither positively nor negatively selected. The dashed vertical line in this specific pathway indicates the nuclear membrane.