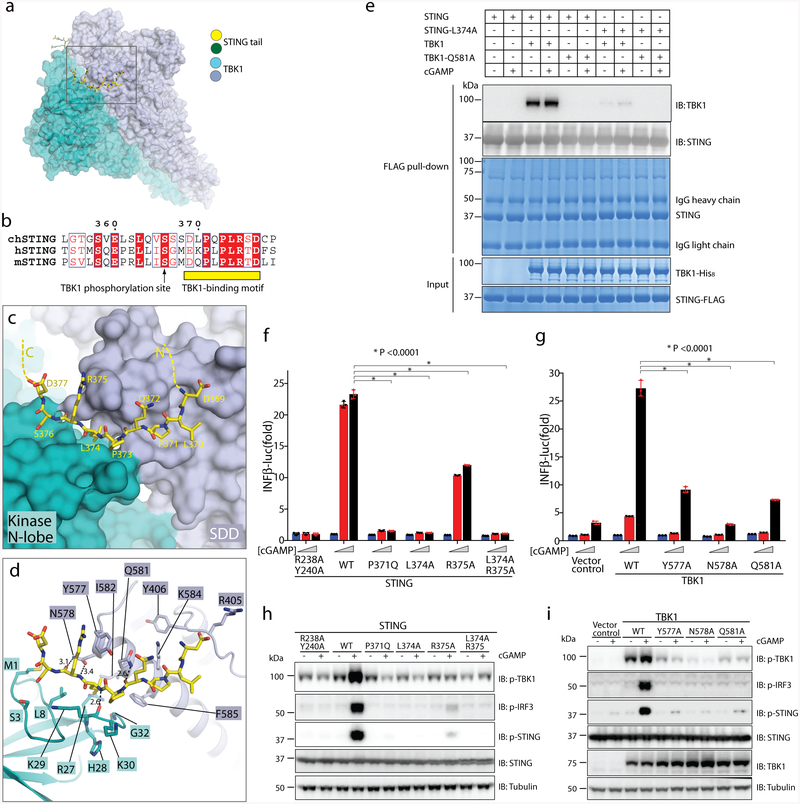
Fig. 2 |. Binding interface between human TBK1 and the chicken STING C-terminal tail.
a, Overview of the binding interface. The rectangle denotes the region shown in detail in c and d. b, Sequence alignment of the C-terminal tail of STING from chicken, human and mouse (denote by ch-, h- and m-prefixes, respectively). c, d, Detailed views of the binding interface. Potential hydrogen bonds are indicated with dotted lines. Numbers are inter-atom distances. N-lobe, N-terminal lobe. e, Mutants of interface residues in either STING or TBK1 abolish the STING/TBK1 interaction in vitro. Data are representative of two independent biological experiments. f, Mutants of interface residues in human STING abolish cGAMP-stimulated IFNβ expression. HEK293T cells that stably express IFNβ–luciferase (IFNβ–luc) were transfected with the STING–Flag wild type (WT) or mutants. The R238A/Y240A mutant (which does not bind cGAMP) served as a negative control. Cells were stimulated with increasing concentrations of cGAMP (0, 0.3 and 1.4 μM). Data are mean ± s.d. n = 3. g, Mutants of interface residues in human TBK1 abolish cGAMP-stimulated IFNβ expression. HEK293T TBK1-null cells that stably express human STING–Flag were reconstituted with the TBK1 wild type or mutants, using lentiviral infection. Cells were transiently transfected with the IFNβ–luciferase reporter and stimulated with increasing concentrations of cGAMP (0, 0.3 and 1.4 μM). Luciferase activity was determined 24 h after transfection. Data are mean ± s.d. h, i, Mutants of interface residues in either STING (h) or TBK1 (i) abolish cGAMP-stimulated phosphorylation (p) of TBK1, STING and IRF3. Cells used in h and i are similar to those in f and g, respectively, except the cells in h and i do not have the IFNβ–luciferase reporter. Cells were treated with cGAMP (1 μM) for 3 h, and subjected to immunoblotting analyses. Data in f–i are representative of three independent biological replicates.