Fig. 1.
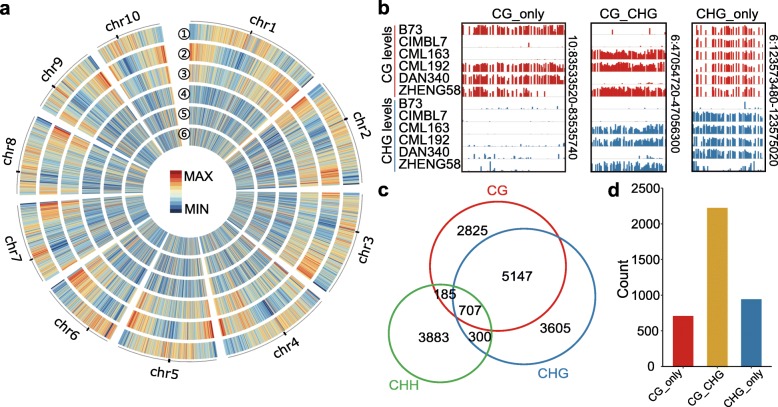
Natural variation of DNA methylation among maize inbred lines. a The distribution of DMRs throughout maize chromosomes and the relationship with other genomic features is visualized. From the outer ring to the inner ring (1–6) are TEs, genes, regions with capture probes, CG DMRs, CHG DMRs, and CHH DMRs. Density of each type was calculated based on 1 Mb windows, and the centromeres are represented by black blocks. b Examples of context-specific DMRs. Each track represents different inbred lines. c Overlaps between CG, CHG, and CHH DMRs. d Total number of context-specific DMRs