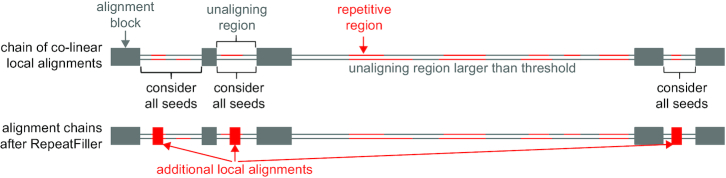
Figure 1:
Missed repeat-overlapping alignments and concept of RepeatFiller. Illustration of RepeatFiller. Focusing on unaligning regions in a reference and query genome that are flanked by up- and downstream aligning blocks, RepeatFiller performs a second round of local alignment considering also repeat-overlapping seeds. Newly found local alignments (red boxes) are inserted into the context of other aligning blocks (grey boxes). Unaligning regions that are larger than a user-defined threshold are not considered because the chance of aligning non-orthologous repeats is increased.