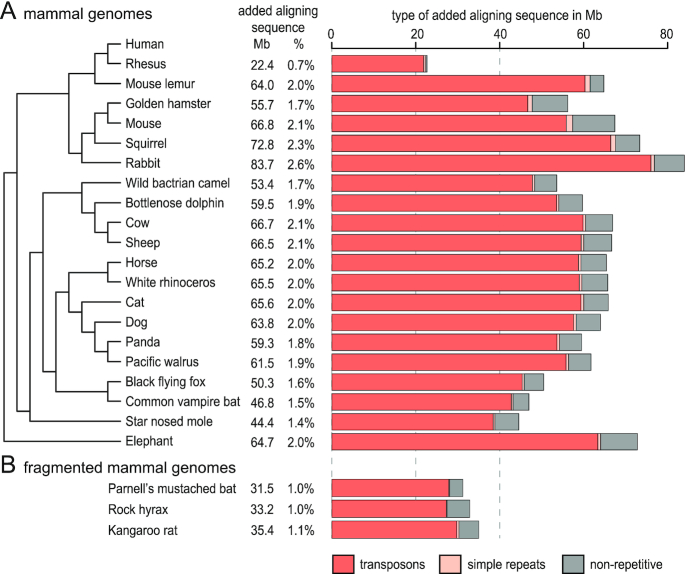
Figure 2:
RepeatFiller adds several megabases of aligning transposable elements to existing mammalian genome alignments. (A) Phylogenetic tree of human and 20 non-human mammals whose genomes we aligned to the human genome. The amount of new alignments detected by RepeatFiller is shown in megabases and in percent relative to the human genome. Bar charts provide a breakdown of newly added aligning sequences into overlap with transposons, simple repeats, and non-repetitive sequence. (B) Application of RepeatFiller to fragmented mammalian assemblies still adds a substantial amount of new alignments.