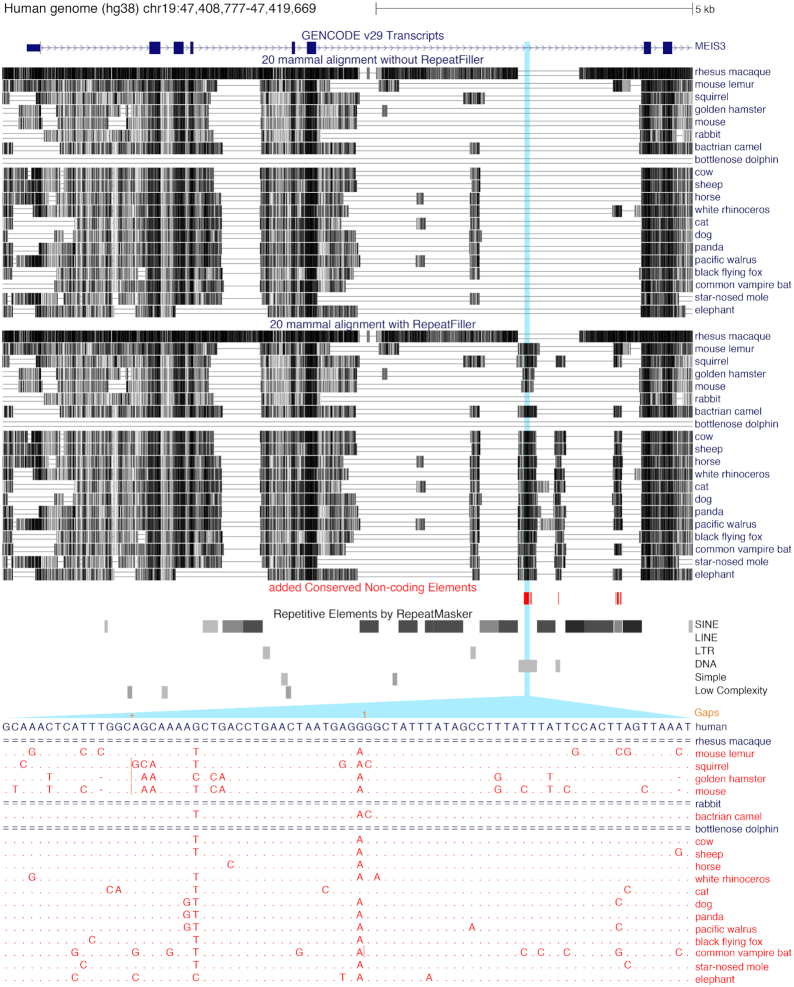
Figure 4:
Examples of newly identified CNEs near MEIS3. UCSC genome browser [45] screenshot shows an ∼11 kb genomic region overlapping the gene MEIS3, a homeobox transcription factor that is required for hindbrain development. Visualization of the 2 multiple genome alignments (without RepeatFiller at the top, with RepeatFiller below; boxes representing align regions with darker colors indicating a higher alignment identity) shows that RepeatFiller adds several aligning sequences, some of which evolve under evolutionary constraint and thus are CNEs (red boxes) only detected in the RepeatFiller-subjected alignment. The RepeatMasker annotation shows that these newly identified CNEs overlap transposons. The zoom-in shows the 21-mammal alignment of one of the newly identified CNEs, which overlaps a DNA transposon. While this genomic region did not align to any mammal before applying RepeatFiller, our tool identified a well-aligning sequence for 17 non-human mammals (red font). A dot represents a base that is identical to the human base, insertions are marked by vertical orange lines, and unaligning regions are showed as double lines.