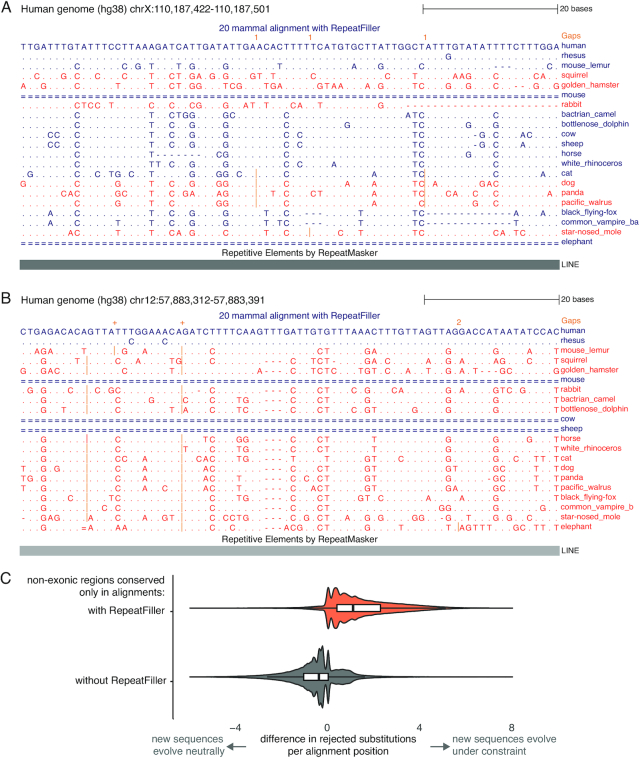
Figure 6:
Additional alignments found with RepeatFiller reveal absence of conservation in the genomic regions that were erroneously classified as conserved before. (A, B) UCSC genome browser screenshots showing 2 examples of genomic regions that were only classified as constrained in a multiple genome alignment generated without applying RepeatFiller. Dots in these alignments represent bases that are identical to the human base, insertions are marked by vertical orange lines, and unaligning regions are shown as double lines. The alignments show that the sequences of species added by RepeatFiller (red font) exhibit a number of substitutions. This explains why these regions were not classified as constrained anymore, despite adding more aligning sequences. Note that in (B) only the sequence of the rhesus macaque was aligned before applying RepeatFiller. Sequences in both (A) and (B) overlap long interspersed nuclear element transposons (LINEs). (C) Difference in evolutionary constraint in non-exonic alignment columns that are only classified as constrained in either alignment. For each alignment position, we used GERP++ to compute the estimated number of substitutions rejected by purifying selection (RS). The difference in RS between alignments with and without RepeatFiller is visualized as a violin plot overlaid with a white box plot (box spans the first to third quartile and indicates the median). This shows that almost all non-exonic bases that were only detected as constrained in the alignment with RepeatFiller (orange background) have a positive RS difference, indicating that the newly aligning sequences added by RepeatFiller largely evolve under evolutionary constraint. In contrast, non-exonic bases only detected as constrained in the alignment without RepeatFiller (grey background) often have slightly negative RS differences, indicating that many of the newly added sequences do not evolve under constraint. The 2 distributions are significantly different (P < E−16, 2-sided Wilcoxon rank sum test).