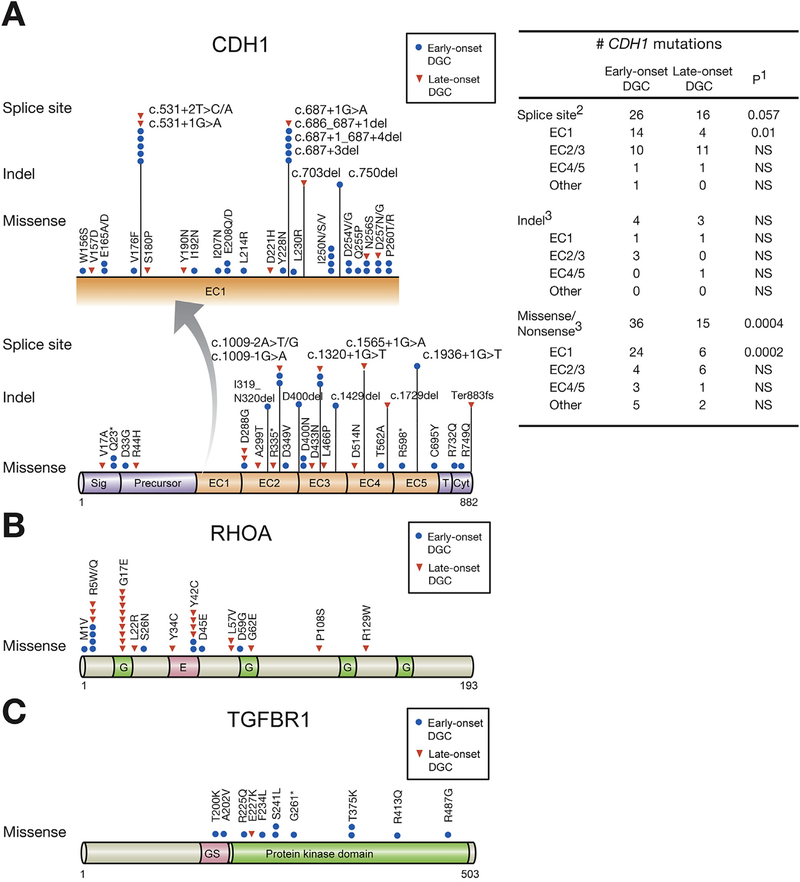
Figure 3.
Somatic mutations in CDH1, RHOA, and TGFBR1 of early- and LODGCs as identified by targeted sequencing. (A) CDH1 mutation sites. Left, CDH1 mutations in the E-cadherin EC1 domain, which are displayed in expanded view (arrow), were more frequent in EODGC (blue circle) than in LODGC (red triangle). “Hot spot” splice site mutations occurred. c.531+1G>A, and c.531+2T>C/A mutations led to exon 4 truncations, and c.687+1G>A, c.686_687+1del, c.687+1_687+4del, and c.687+3del mutations resulted in truncation of exon 5 in EC1 domain. Right, the number of somatic CDH1 mutation sites in EODGCs (n = 109) and LODGCs (n = 115) according to the targeted sequencing; 1P value by χ2 test. 2The number of CDH1 splicing aberrations that were identified either by targeted DNA sequencing or by reverse transcription polymerase chain reaction. 3The number of CDH1 mutations that were identified by targeted DNA sequencing (B) RHOA mutation sites (C) TGFBR1 mutation sites.